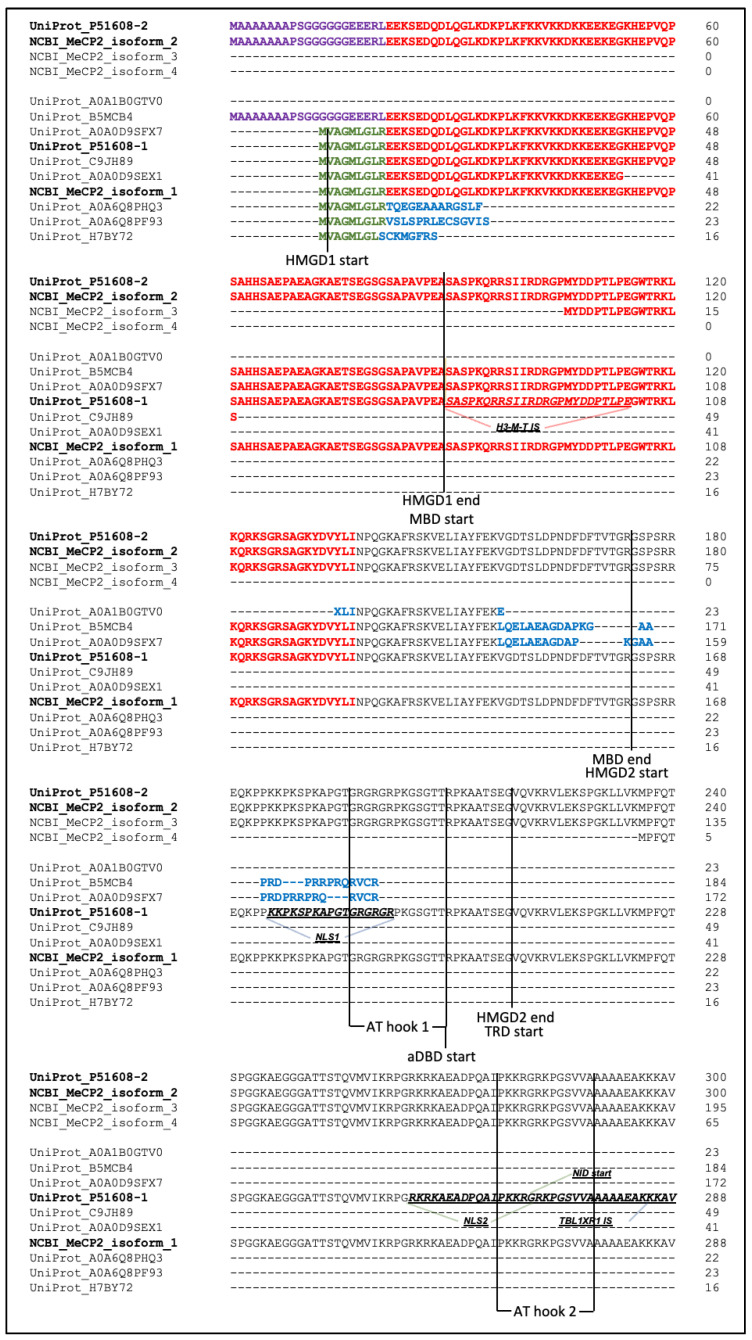
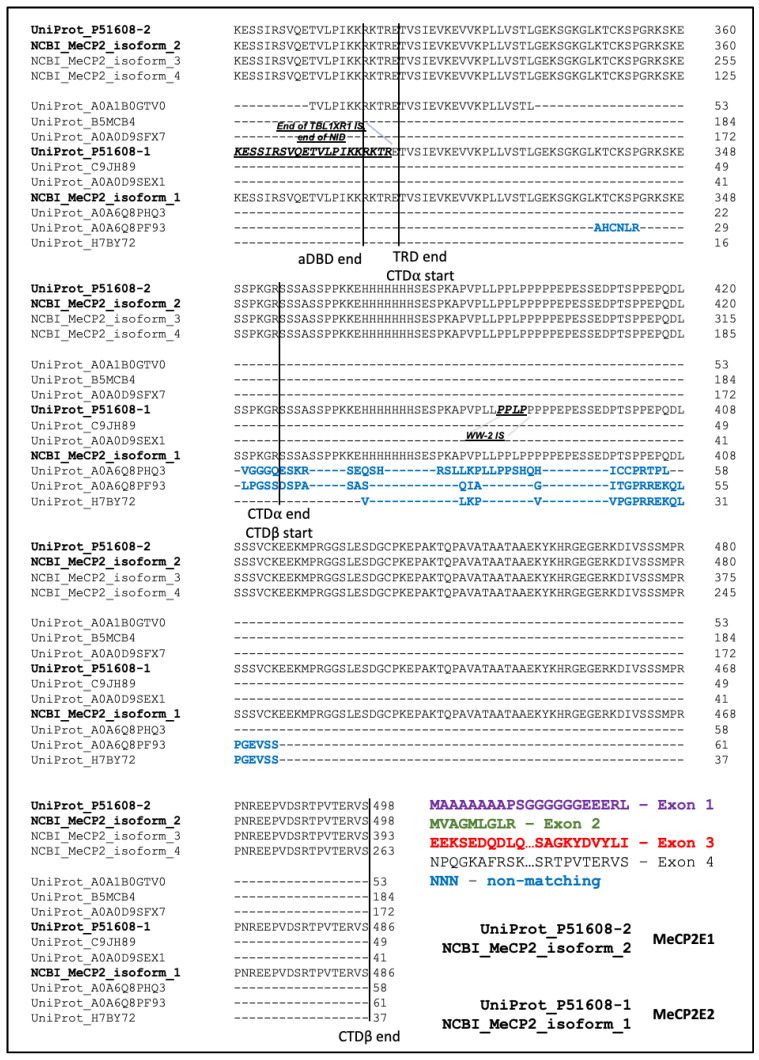
Figure 1.
Alignment of potential MeCP2 protein isoforms from NCBI and UniProt. The sequences are in the N-terminal to C-terminal orientation. MeCP2E1 and MeCP2E2 from both NCBI and UniProt are shown in bold. The raw output from ClustalOmega was split into 2 panes and exons were colour-coded as indicated in the figure legend. Functional domains are labeled with start and end flags on MeCP2E2 (NCBI isoform 1 and UniProt P51608-1), while protein interaction sequences were underlined. The sequences of exons 1, 2, 3, and 4 were coloured purple, green, red and black respectively. Non-matching to either isoform sequences were coloured blue. aDBD: alternative DNA binding domain; CTDα: C-terminal domain α; CTDβ: C-terminal domain β; HMGD1: high mobility group protein-like domain 1; HMGD2: high mobility group protein-like domain 2; MBD: methyl-CpG-binding domain; NID: NCoR/SMRT interaction domain; NLS1: nuclear localization signals 1; NLS2: nuclear localization signals 2; TBL1XR1 IS: TBL1XR1 interaction sequence; TRD: transcriptional repression domain; WW-2 IS: Group 2 WW motif-containing protein interaction sequence. References and amino acid numbers of each feature were summarized in Supplementary Table S1.