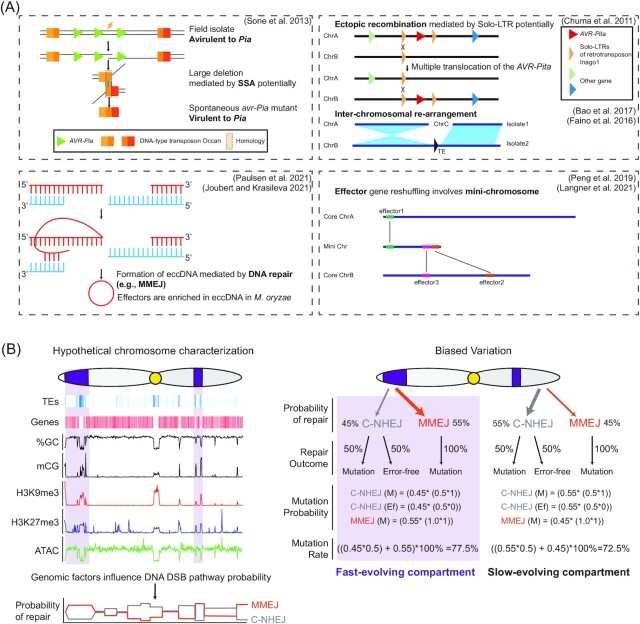
Figure 4.
The association among DSB repair, genome compartments and biased genome evolution in filamentous pathogens. (A) Examples of observed genome variation influenced by DNA repair. Transposable element (TE) or other repeated DNA can mediate ectopic recombination or re-arrangements (top). DNA contained outside of the core genome, such as eccDNA and mini- or dispensable-chromosomes, may arise due to DNA repair mechanisms, or shuffle DNA with core chromosomes, contributing to biased genome variation in filamentous pathogens (bottom). (B) Illustration of a hypothetical chromosome characterized for multiple genomic, epigenomic and chromatin variables. These factors may collectively influence the DNA repair hierarchy at a given genomic locus. We provide a specific example of how DNA repair choice between C-NHEJ and MMEJ could lead to different rates of DNA mutation in the genome, a model we refer to as Biased Variation. Note, half of the DNA DSBs repaired by C-NHEJ may not cause any DNA mutation, which we represent as 0 in the mutation probability calculation, while half and all DNA DSBs repaired by C-NHEJ and MMEJ will results in a DNA mutation, represented by a 1 in the mutation probability calculation. The rates and frequencies used here are approximations and remain to be experimentally determined. (M), mutation; (Ef), error-free.