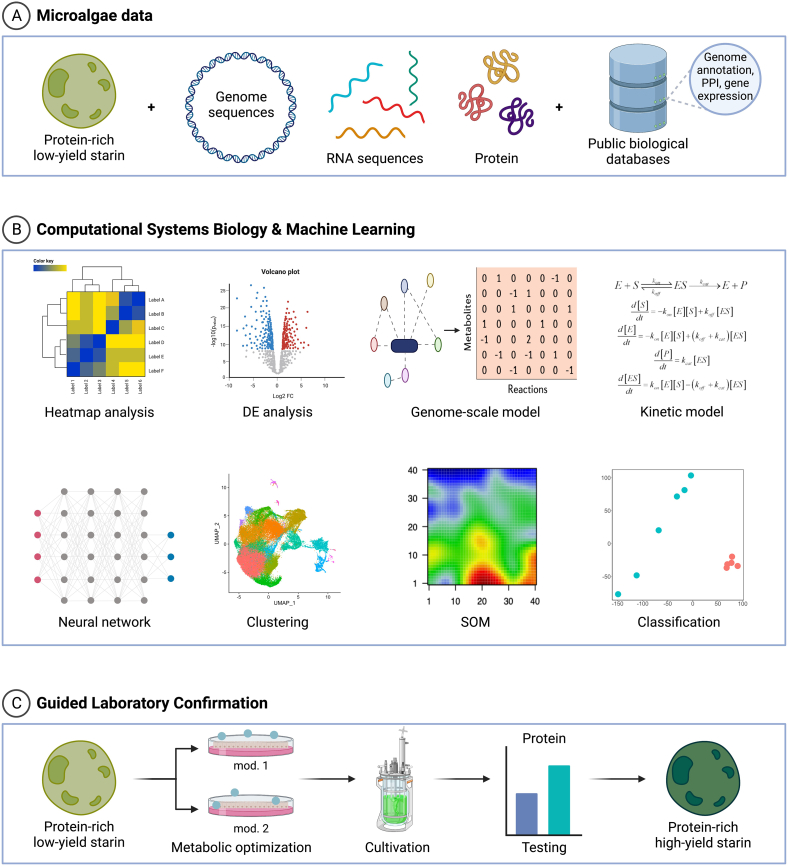
FIGURE 1.
Systems biology and machine learning pipeline. (A) Microalgae data can be stored or retrieved from private/public databases. They can contain gene/protein sequences and structures and omics data (transcriptomics, proteomics, and metabolomics). (B) The data from (A) can be used to perform biostatistics, data analytics, and machine learning to identify expression correlations; co-regulated clusters; and differentially activated genes, proteins, or metabolites between samples. Metabolites and enzyme data can be used to develop and test a genome-scale model to identify growth constraints, as well as kinetic models, which could highlight bottlenecks in metabolic fluxes. (C) The model-optimized microalga strains can then be experimentally tested for better yield, especially at a commercial scale. DE, deferential expression; SOM, self-organization map.