Figure 2. Lettuce likely contains a G-quadruplex in its DFHBI-1T-binding region.
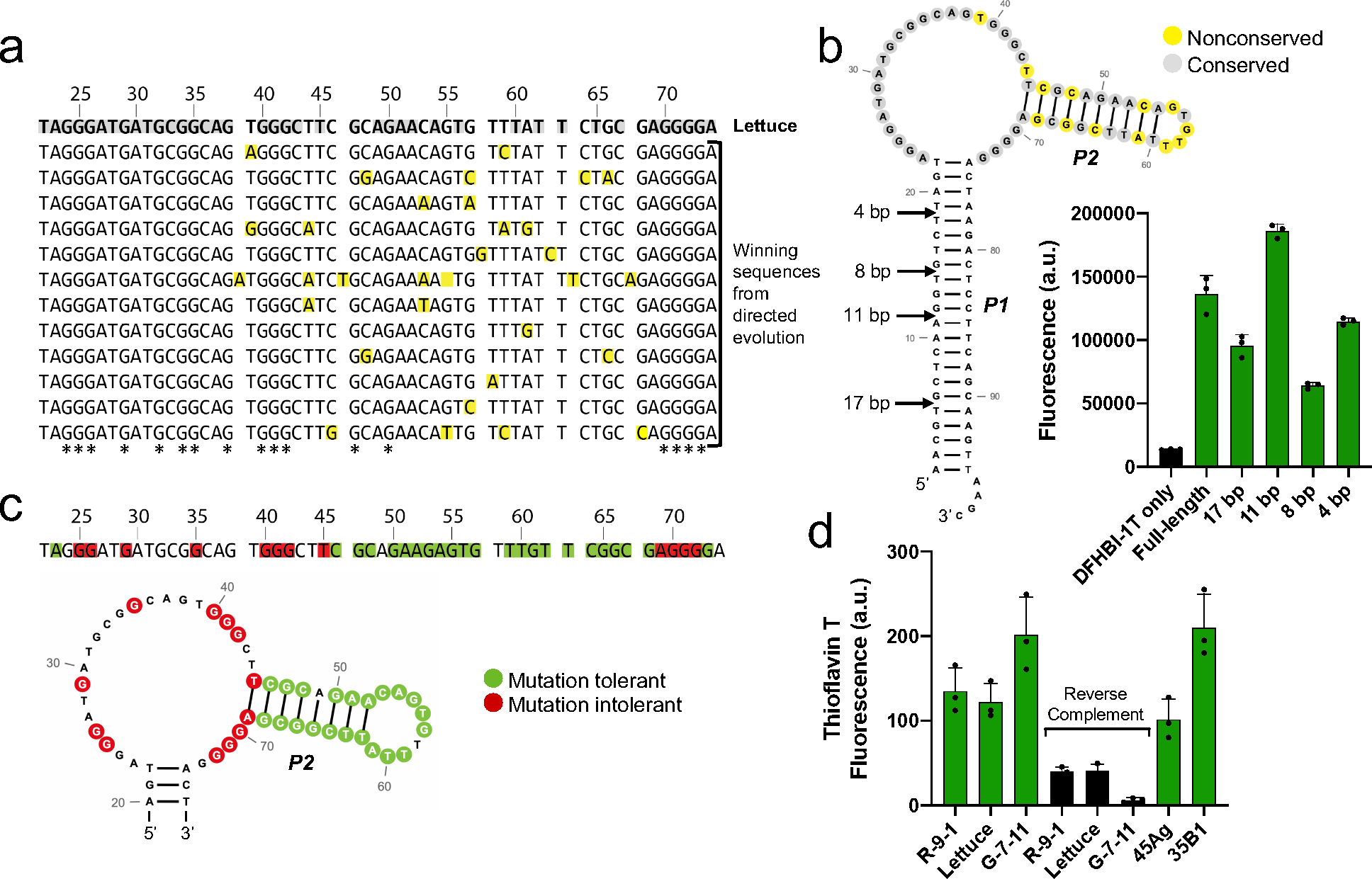
a) Sequence alignment of aptamers that retained the ability to induce DFHBI-1T fluorescence after directed evolution of Lettuce. All aptamers exhibited similar fluorescence signal to that of Lettuce. The top sequence is Lettuce, and all sequences below are sequences of functionally active aptamers discovered from directed evolution of Lettuce. Yellow-shaded nucleotides differ from the sequence of Lettuce. Grey-shaded nucleotides in the Lettuce sequence were conserved across all functionally active aptamers from the direct evolution experiment. G residues that are conserved across all functionally active aptamers are indicated in the bottom row with an asterisk. Fluorescence emission was measured at (excitation 460 nm, emission 480–560 nm) for all mutants using 10 μM DNA and 20 μM DFHBI-1T. b) Lettuce’s long stem is nonessential for fluorescence activation of DFHBI-1T. Arrows indicate the lengths to which the stem was truncated. Fluorescence was measured (excitation 460 nm, emission 505 nm) of each truncated construct using 1 μM DNA and 5 μM DFHBI-1T. The secondary structure prediction of Lettuce is color coded as in Figure 2a, where yellow represents nonconserved positions and grey represents conserved positions, c) Summary of mutational analysis. To identify essential and nonessential residues for fluorescence, Lettuce was mutated at single positions and the fluorescence of each mutant was assessed, where mutants that retained fluorescence within 10% of Lettuce were considered tolerant, and mutants with fluorescence values below 30% of Lettuce were considered to be intolerant. Green and red shaded nucleotides indicate positions at which a mutation was tolerated or not tolerated, respectively. Unshaded positions were not tested. Fluorescence emission was measured (excitation 460 nm, emission 480–560 nm) for 35 mutants of Lettuce using 10 μM DNA and 20 μM DFHBI-1T. d) Fluorescence activation of thioflavin T by DNA aptamers. R-9-1, Lettuce, and G-7-11 were incubated with thioflavin T, a compound known to bind G-quadruplexes. Reverse complement sequences which are not expected to form G-quadruplexes were tested, along with two sequences known to form G-quadruplexes (45Ag and 35B1). Fluorescence (excitation 430 nm, emission 485 nm) was measured using 1 μM DNA and 1 μM thioflavin T.
