Figure 5. Optimization of split Lettuce for detection of target RNA.
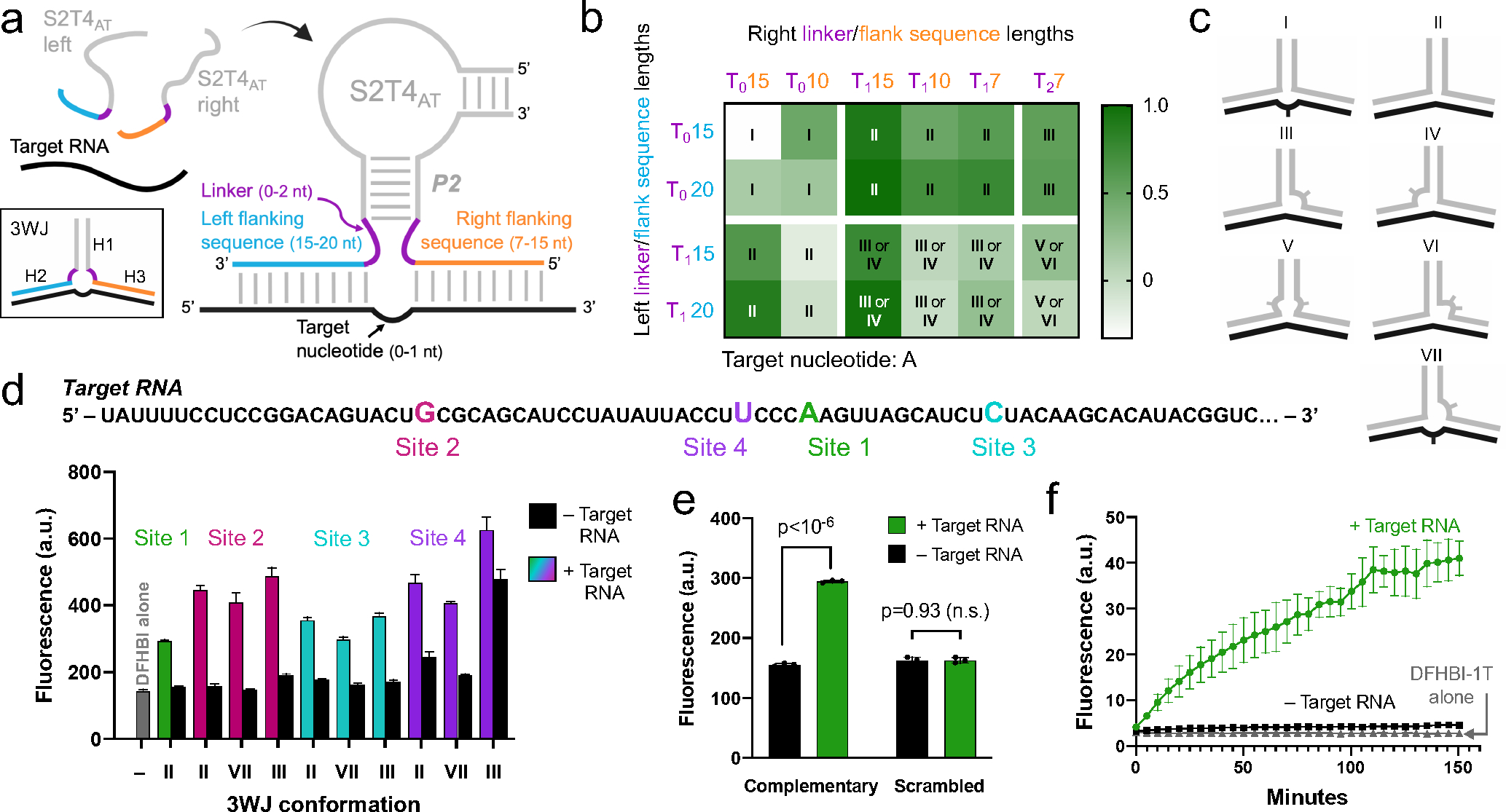
a) Diagram of the split Lettuce RNA sensor and regions to be optimized. Each half of S2T4at can assemble on a target RNA sequence via flanking sequences (orange and blue) that are complementary to the target RNA (black), forming a three-way junction (3WJ) between the two Lettuce oligonucleotides and the target RNA. The three helices (H1-H3) of the 3WJ are indicated. The left and right strands we tested had 7, 10, 15, or 20-nt flanking sequences and 0, 1, or 2 linker residues (purple). We tested RNA target sites with either one or zero unpaired target nucleotides, b) Sensor pairs have various 3WJ conformations and activate a range of fluorescence signal. Left and right strands with the indicated lengths of thymine linkers (purple) and flanking sequences (blue and orange) were designed to target a single site on a target RNA. Fluorescence of each pair was measured with and without target RNA. Heat map values were obtained by subtracting the fluorescence of samples without target RNA from samples with target RNA, and values over 0.5 were considered successful. Roman numerals refer to each pair’s 3WJ structure, which are shown in Figure 5c. The 3WJ assignments take into account that the target nucleotide, adenine, can base pair with thymines in the linkers. Type II and III 3WJs were the most successful on this particular target site (Figure 5d, Site 1). c) Diagrams of a selection of the 3WJs that can form between target RNA (black) and the two Lettuce sensor strands (grey). The location of unpaired nucleotides is indicated by curves, where one and two tick marks correspond to one and two unpaired nucleotides, d) Sensor designs that were successful on one target RNA site also work on three other target RNA sites. Sensor pairs of type II, type III and type VII 3WJs were designed to target three other RNA sites (Site 2, Site 3, and Site 4) which each have a different target nucleotide (G, C, and U, respectively). For comparison, the highest-performing sensor from Figure 1b, which targets Site 1, is included. The majority of the sensors exhibited signal-to-background ratios as high or higher than the original Site 1 sensor, showing that these sensor designs can be generalized to detect other RNA targets of interest. Fluorescence was measured (excitation 460 nm, emission 505 nm) using 10 μM DNA, 5 μM DFHBI, and 5 μM RNA on a Gen5 fluorescence plate reader. Error bars indicate s.d. (n=3). e) Split Lettuce RNA detection is sequence-dependent. To confirm the specific base pairing between flanking sequences and the target RNA sequence, we randomly scrambled the flanking sequences of the Site 1 sensor shown in Figure 5d. No increase in fluorescence is observed when target RNA is present. With fully complementary flanking sequences, the sensor exhibits an increase in fluorescence when target RNA is present. Fluorescence was measured as described in Figure 5d. Error bars indicate s.d. (n=3). f) Time dependence of split Lettuce sensor fluorescence. Split Lettuce halves and DFHBI-1T were incubated with or without target RNA and fluorescence was measured as described in Figure 5d at different time points for a total of 150 minutes. Error bars indicate s.d. (n=3).
