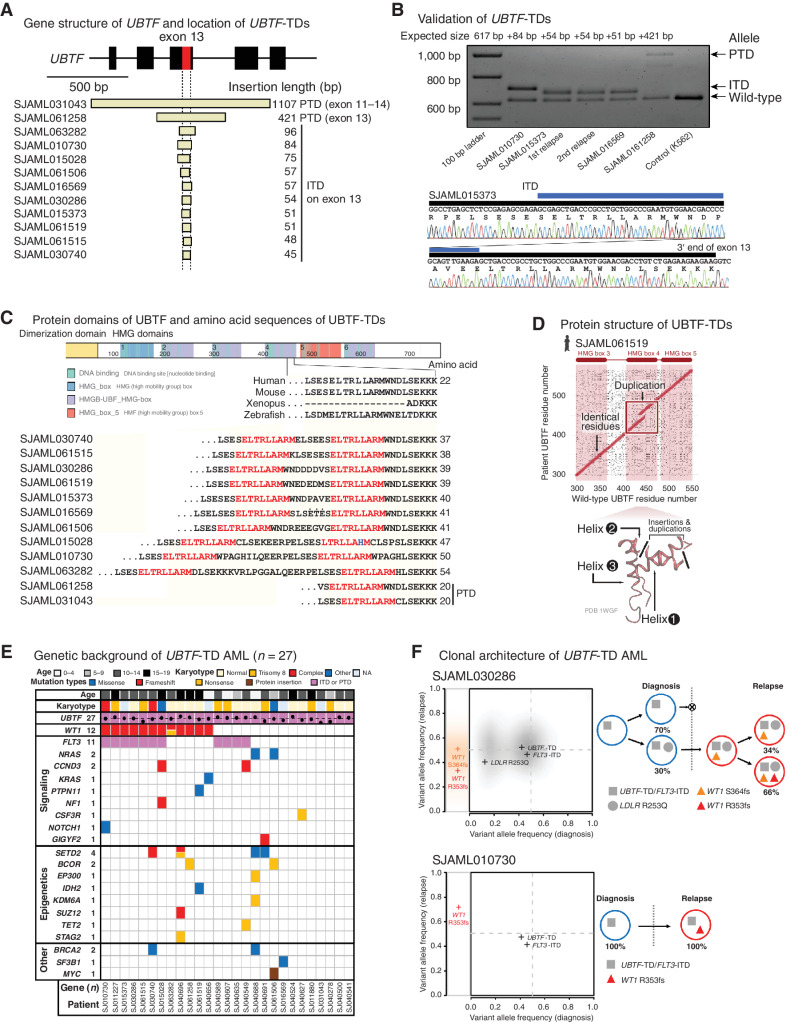
Figure 2.
UBTF-TDs in pediatric AML. A, Exons 11–15 of UBTF gene and the location of UBTF-TDs (n = 12) identified in the relapse AML cohort. B, The results of validation of UBTF-TDs in the relapsed AML cohort by polymerase chain reaction (PCR) and Sanger sequencing. Blue bars denote duplicated sequences. C, Illustrative schema of UBTF protein and amino acid sequences within the HMG domain 4 of both UBTF-wild-type and UBTF-TDs. A part of UBTF-TDs encoded on exon 13 of UBTF genes is shown in comparison with UBTF-wild-type of human and other vertebrates. Amino acid sequences highlighted in red denote leucine-rich sequences duplicated in all UBTF-TDs. D, Comparisons of amino acid sequences of UBTF-wild-type and UBTF-TD at the likely insertion site in helix 2 of HMB box 4 for observed UBTF-TDs. E, Mutational landscape of UBTF-TD AML. F, Clonal dynamics of WT1 mutations in UBTF-TD AMLs. Comparison of variant allele frequency between diagnosis (x-axis) and relapse (y-axis) tumors for cases SJAML030286 and SJAML010730 (left). SNVs/Indels detected from SJAML030286 WGS were drawn as density clouds, and representative mutations for each subclone were marked by crosses. Relapse-specific mutations are shown to the left. The clonal evolution scheme for the patients imputed from bulk WGS data (SJAML030286) or RNA-seq and TCS (SJAML010730; right).