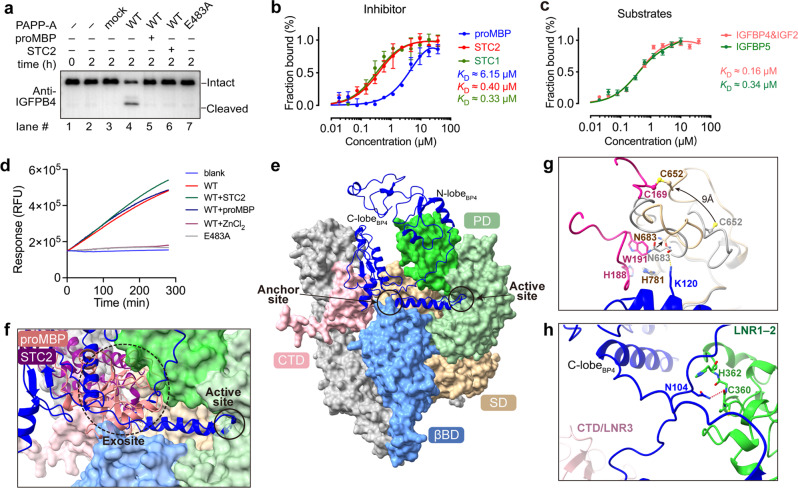
Fig. 5. Proteolytic inhibition of PAPP-A activity requires the exosite binding of proMBP or STC2.
a Cleavage of the full-length IGFBP4 in the presence of IGF-2 was assessed using in vitro reaction. The bands of intact and cleaved IGFBP4 detected by western blot analysis are indicated. Reactions with empty vector (mock) and inactive PAPP-A (E483A) were used as negative controls. b Microscale thermophoresis (MST) analyses of proMBP, STC1, and STC2 bound to PAPP-A. The identical batch of PAPP-A protein was used. The signals were consonant with the bound fraction, generating a dissociation constant (KD) that was measured from three biologically independent repeats (n = 3). c MST analyses of substrates bound to PAPP-A. KD was measured from three biologically independent repeats (n = 3). d Fluorescence resonance energy transfer (FRET) analyses of PAPP-A cleavage on 4P1 under different inhibitory conditions. The absence of PAPP-A (blank), inactive PAPP-A (E483A), and the addition of ZnCl2 (WT + ZnCl2) were used as negative controls. e AlphaFold-predicted IGFBP4·PAPP-A complex model with IGFBP4 shown in cartoon (blue) and PAPP-A shown in surface (color-coded). f ProMBP or STC2 competes with the substrate for binding PAPP-A at the exosite. g Close-up view of proMBP and IGFBP4 binding on PAPP-A. SD is shown either in tan (in the PAPP-A·proMBP complex) or gray (in the predicted model). h In the predicted IGFBP4·PAPP-A structure, His362 of LNR1–2 inserts into a groove formed by a linker between the N-lobe and the anchor peptide, and Asn104 of IGFBP4 forms a hydrogen bond with the main chain of Cys360 of LNR1–2.