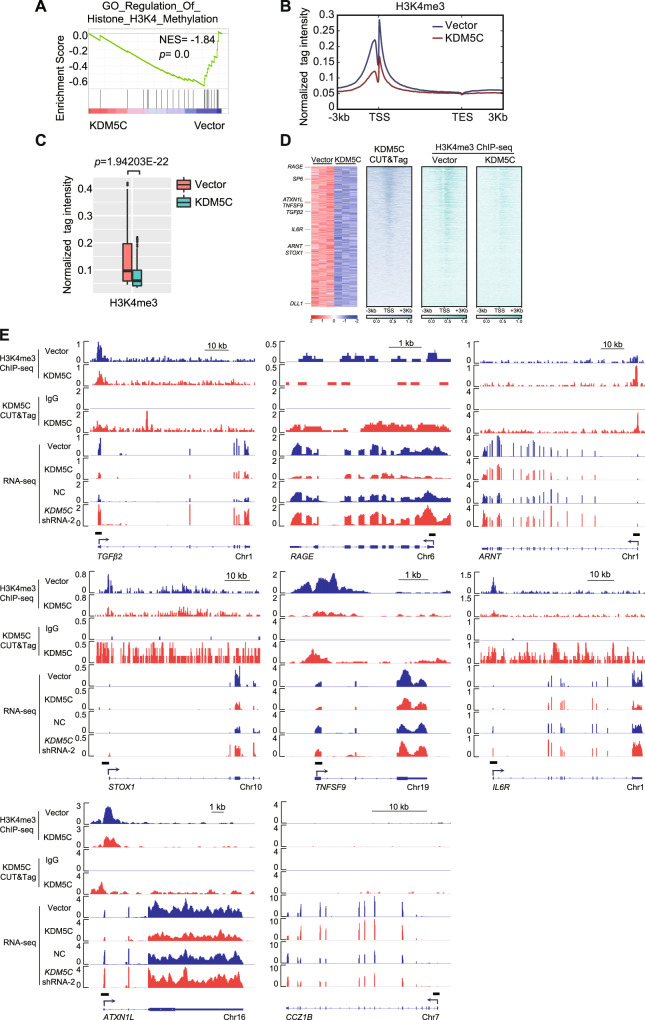
Fig. 5. KDM5C regulates dynamic H3K4me3 in the promoters of target genes.
A GSEA of the expression profile of HTR-8 cells that were stably transduced with either control vector or KDM5C overexpression vector via a histone H3K4 methylation-associated signature. B Pattern of H3K4me3 signal profile within ±3 kb genomic regions from anchors on the transcriptional start site (TSS) and transcriptional termination site (TTS) of HTR-8 cells stably transduced with either control vector or KDM5C overexpression vector. C Box plots indicating the changes in H3K4me3 occupancy around promoters (within 3 kb genomic regions flanking the TSS) of HTR-8 cells stably transduced with either control vector or KDM5C overexpression vector. D Heatmap representation of the downregulated genes in HTR-8 cells under KDM5C overexpression conditions. The binding profiles of KDM5C or H3K4me3 within 3 kb genomic regions flanking the TSS of each gene are also shown. E H3K4me3 ChIP-seq in HTR-8 cells stably transduced with either control vector or KDM5C overexpression vector, IgG or KDM5C CUT&Tag in HTR-8 cells stably transduced with KDM5C overexpression vector, and RNA-seq of HTR-8 cells stably transduced with either control vector or KDM5C overexpression vector. Genome browser tracks representing sites for H3K4me3 binding or KDM5C at TGFβ2, RAGE, ARNT, STOX1, TNFSF9, IL6R, ATXN1L, or CCZ1B gene loci.