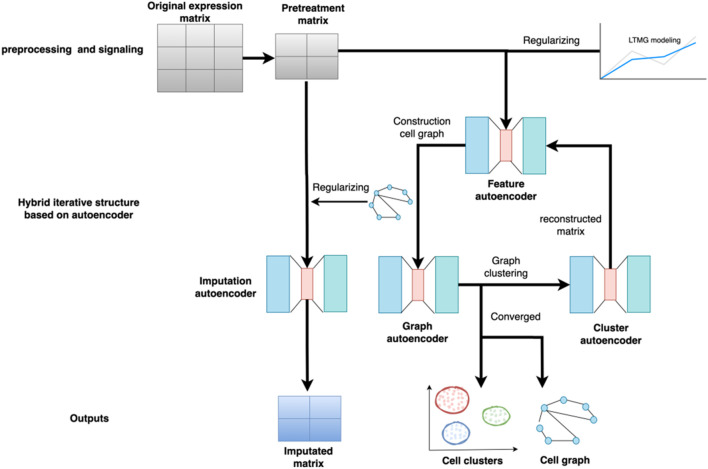
FIGURE 1.
Structure of the scGAEGAT model. The model uses the pretreatment gene expression matrix as input to the feature autoencoder. The feature autoencoder builds and prunes the cell graph through learned embedding. The graph autoencoder adds the graph attention mechanism. It takes the constructed cell graph as input by adding different weights to different nodes, which can better capture cell relationships and achieve cell-type clustering. Each type of cell has a separate cluster autoencoder to reconstruct the gene expression value of the cell. The reconstructed gene expression value is used as the new input for iteration until convergence.