Table 5.
The abnormal expressions of key genes constituting the prognostic model.
| Gene | Feature | Remark | Reference |
|---|---|---|---|
| CDKN2A |

|
CDKN2A is also known as P16. Yoon et al. illustrated that the expression of p16 was up-regulated in TNBC sample by using tissue microarray, and a significant co-expression correlation existed between p16 and SRY-box transcription factor 10 (SOX10) but not in non-TNBC cases. | (15) |
| PSAT1 |

|
Metcalf et al. showed that the expression of PSAT1 was up-regulated in TNBC by using cell and qPCR, and the expression of PSAT1 increased with the increase of the clinical grade of TNBC. | (16) |
| HMGB1 |

|
Lee et al. found that HMGB1 was highly expression in TNBC by using patient tissues and immunohistochemistry, and the expression of HMGB1 was notably correlated with tissue grade, abundant tumor infiltrating lymphocytes and CD8 positive cells. | (17) |
| ELAVL1 |

|
ELAVL1 is also known as HuR. Mehta et al. verified that HuR was up-regulated in TNBC cells compared with normal cells using cellular and western-blot analysis, and knockdown of HuR induced oxidative stress and DNA damage in cells, and further enhanced sensitivity of TNBC cells to radiotherapy. | (18) |
| SLC7A11 |

|
Bai et al. illustrated that the expression of SLC7A11 was up-regulated in TNBC cells compared with normal cells by immunohistochemistry test and the overexpression of SLC7A11 was connected with the promotion of glutathione biosynthesis, which further contributed to a radioresistance in TNBC. | (19) |
| ASNS |

|
Qin et al. showed that ASNS highly expressed in breast cancer by using cell and qPCR, and the high expression of ASNS was correlated with poor prognosis. | (20) |
| LAMP2 |

|
Damaghi et al. illustrated that LAMP2 highly expressed in breast cancer by using proteomics and qRT-PCR, which was positively associated with the course of disease. | (21) |
| CAV1 |

|
Witkiewicz et al. found that down-regulated CAV1 in breast cancer was correlated to poorer overall survival by using patient tissues and immunostaining. | (22) |
| HELLS |

|
Hou et al. showed that HELLS was up-regulated in pancreatic tissues and correlated with advanced clinical stage and poor prognostic by using qRT-PCR. Knockdown of HELLS could lead to tumor cell arrest and increase sensitivity to cisplatin. | (23) |
| DPP4 |

|
Choy et al. illustrated that DPP4 was notably down-regulated in breast cancer through comparative study using multiple public databases, and the down-expression of DPP4 may be related to the poor prognosis of patients. | (24) |
| TF |

|
Hong et al. found that TF was down-regulated in melanoma by comparing the GSEA databases, and their study showed that the lipidogen regulator Sterol-regulatory element binding proteins (SREBP2) could directly induce the transcription of the TF, thereby decreasing the intracellular iron pool, the activity of oxygen and lipid peroxidation, and inhibiting ferroptosis in cells. | (25) |
| ZFP69B |
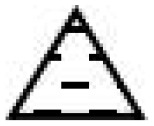
|
Zheng et al. showed that the expression of ZFP69B was up-regulated in gastric cancer by data mining through TCGA and GEO database, and a prognostic model composed of ZFP69B they further built could well forecast the prognosis of patients. | (26) |
 Gene was confirmed to be up-regulated by experiment in TNBC;
Gene was confirmed to be up-regulated by experiment in TNBC;
 Gene was confirmed to be up-regulated by experiment in breast cancer;
Gene was confirmed to be up-regulated by experiment in breast cancer;
 Gene was confirmed to be down-regulated by experiment in breast cancer;
Gene was confirmed to be down-regulated by experiment in breast cancer;
 Gene was confirmed to be up-regulated by experiment in other cancers;
Gene was confirmed to be up-regulated by experiment in other cancers;
 Gene was confirmed to be down-regulated by calculation in breast cancer;
Gene was confirmed to be down-regulated by calculation in breast cancer;
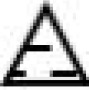 Gene was confirmed to be up-regulated by calculation in other cancers;
Gene was confirmed to be up-regulated by calculation in other cancers;
 Gene was confirmed to be down-regulated by calculation in other cancers。
Gene was confirmed to be down-regulated by calculation in other cancers。
