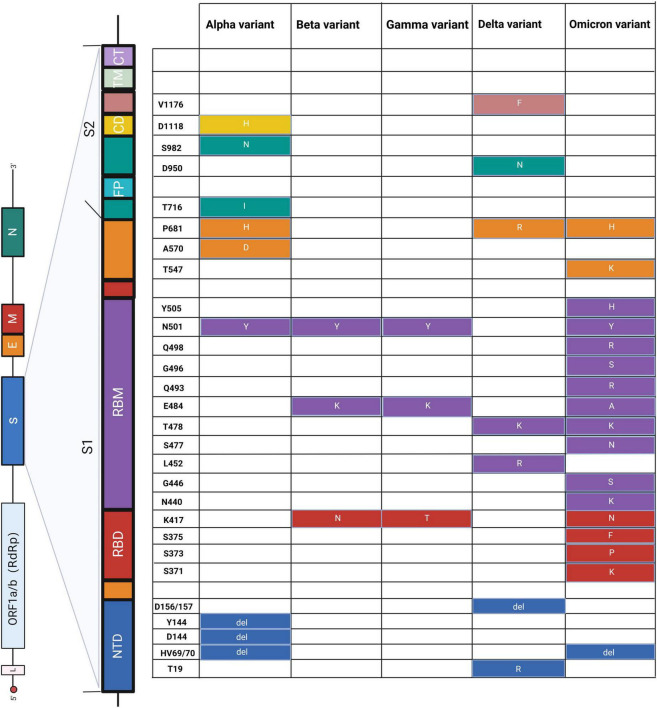
FIGURE 3.
Schematic diagram of amino acid mutations in the S gene of SARS-CoV-2. The left panel shows the genome of SARS-CoV-2 and the detailed structure of the S gene. The S1 and S2 subunits and a transmembrane domain constitute the spike protein (S). RBD and RBM bind to the host cell receptor. S2 consists of FP, CD, and CT, contributing to membrane fusion. Substitutes of amino acids of mutations are presented on the right panel, and the colors correspond to the structure of the S gene. NTD, N-terminal domain; RBD, receptor-binding domain; RBM, receptor-binding motif; FP, fusion peptide; CD, connecting domain; TM, transmembrane domain; CT, cytoplasmic tail. Alpha (Berenger et al., 2021; Borsova et al., 2021; Hale et al., 2021; Fu et al., 2022; Oh et al., 2022; Ratcliff et al., 2022), beta (Hale et al., 2021; Dikdan et al., 2022; Fu et al., 2022), gamma (Hale et al., 2021; Ratcliff et al., 2022), delta (Berenger et al., 2021; Hale et al., 2021; Norz et al., 2021; Oh et al., 2022; Rosato et al., 2022), and omicron (Bloemen et al., 2022; Dachert et al., 2022; Rasmussen et al., 2022).