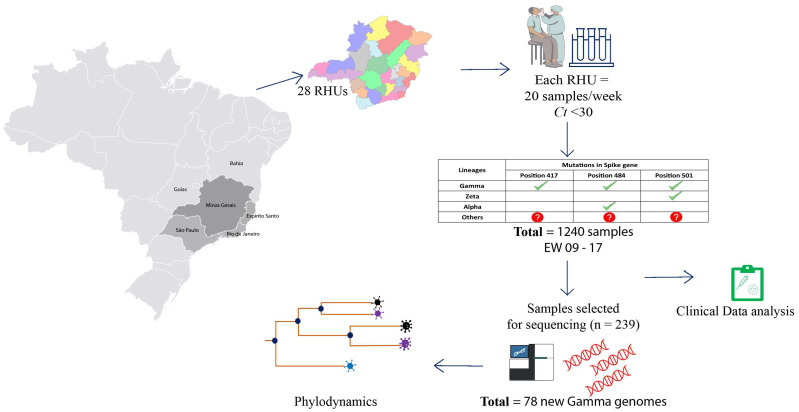
Figure 1.
Workflow strategy for genomic evaluation of SARS-CoV-2 variants in MG. Twenty samples/week positive for SARS-CoV-2 with Ct < 30 were obtained from 28 Regional Health Units. Samples were screened for specific mutations in the Spike gene through PCR-genotyping (K417T, E484K and N501Y). Samples with mutations in each position are marked with green check symbol. Samples that presented different mutational profile are marked with red interrogation symbol. The description of the 1240 samples obtained, collection date, municipality of origin and genotyping result are included in Supplementary Table S1. A subset (239 samples) was selected for genome sequencing and phylogenetic analysis. A total of 78 new Gamma genomes were generated by our study and the genomes were used to infer the probable date of insertion in MG.