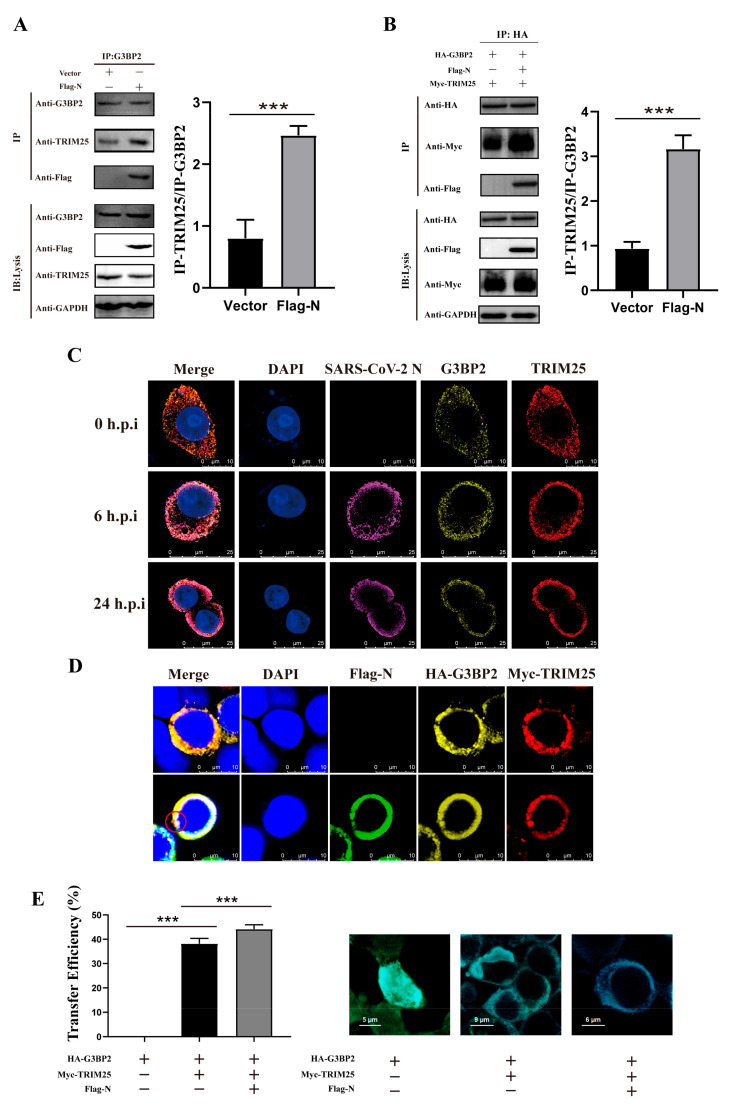
Figure 4.
The engagement of G3BP2-TRIM25 binding is enhanced by the recruitment of the N protein. (A) The pcDNA3.1 vector and pcDNA3.1-N-Flag were transfected into cells. Twenty-four hours after transfection, coimmunoprecipitation showed that binding of TRIM25 to G3BP2 increased significantly in the presence of N. Grayscale results were analyzed using a Gel-Pro analyzer. Data are presented as means ± SD (paired t test, n = three biological replicates per group, * 0.01 < p < 0.05, ** 0.001 < p < 0.01, *** p < 0.001). (B) pcDNA3.1-TRIM25-Myc, pcDNA3.1-G3BP2-HA and pcDNA3.1-N-Flag were cotransfected into cells. Twenty-four hours after transfection, coimmunoprecipitation showed that binding of TRIM25 to G3BP2 increased significantly in the presence of N. Grayscale results were analyzed using a Gel-Pro analyzer. Data are presented as means ± SD (paired t test, n = three biological replicates per group, * 0.01 < p < 0.05, ** 0.001 < p < 0.01, *** p < 0.001). (C) From 0 to 24 h after cells were infected with SARS-CoV-2, confocal imaging showed that TRIM25 and G3BP2 gradually colocalized with N in the cytoplasm from 6 to 24 h.p.i. Red fluorescence corresponds to the TRIM25-positive signal, yellow fluorescence indicates G3BP2, purple fluorescence corresponds to SARS-CoV-2 N, and blue fluorescence corresponds to nuclei (DAPI staining; scale bar, 10 μm). (D) pcDNA3.1-TRIM25-Myc, pcDNA3.1-G3BP2-HA and pcDNA3.1-N-Flag were cotransfected into cells, and confocal imaging showed that TRIM25 and G3BP2 colocalized with N in the cytoplasm. Red fluorescence corresponds to the TRIM25-positive signal, yellow fluorescence labels G3BP2, green fluorescence corresponds to SARS-CoV-2 N, and blue fluorescence corresponds to nuclei (DAPI staining; scale bar, 10 μm). (E) We divided the plasmid into three groups, the first group was pcDNA3.1-G3BP2-HA; the second group was pcDNA3.1-TRIM25-Myc and pcDNA3.1-G3BP2-HA; the third group was pcDNA3.1-TRIM25-Myc, pcDNA3.1-G3BP2-HA and pcDNA3.1-N-Flag was transfected into 293T cells. Twenty-four hours later, the cell samples were collected for immunofluorescence staining. Samples were analyzed by fluorescence resonance energy transfer analysis software. Data are presented as means ± SD (paired t test, n = 5 biological replicates per group, *** p < 0.001).