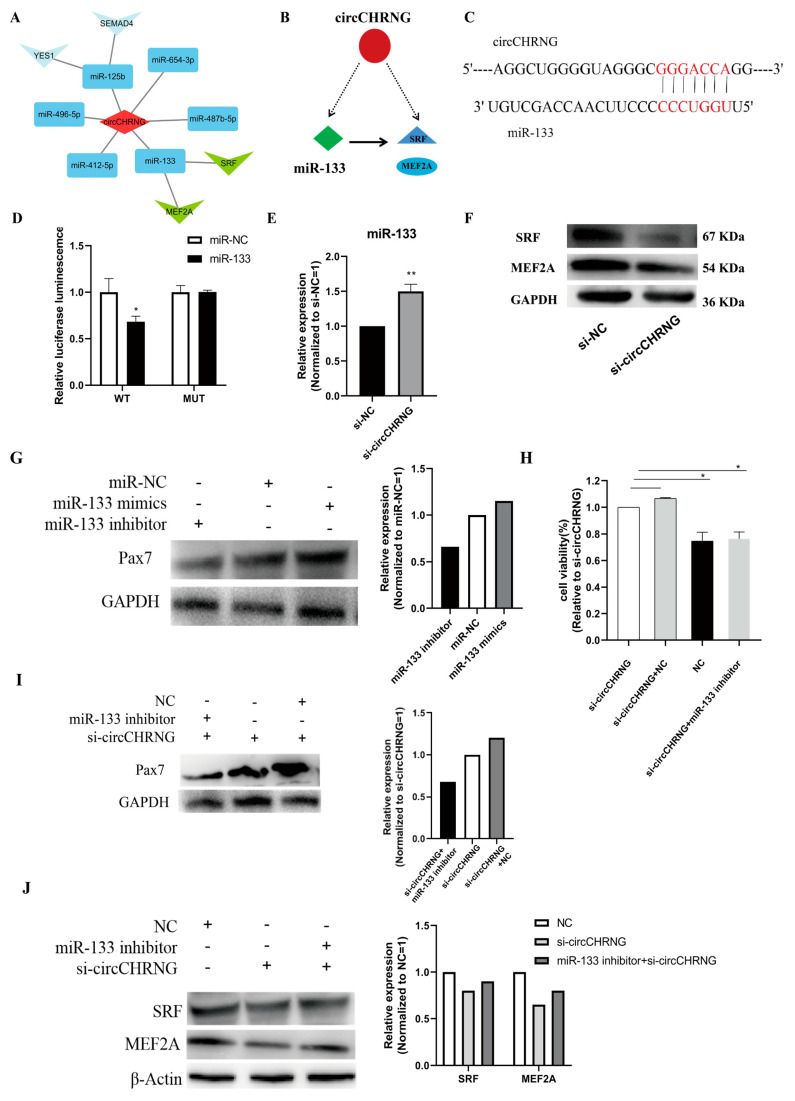
Figure 6.
CircRNA–miRNA–mRNA Regulatory Circuitry. (A) Interaction network of circCHRNG and miRNAs. (B) Illustration of predicted circuitry including miR-133, circCHRNG, and SRF. The red node represents circRNAs; green node represents miRNA; blue node represents key genes related to myogenesis; the solid line represents the real regulatory effect; the dashed line represents the predicted regulatory effect. (C) The potential binding sites of miR-133 in circCHRNG. (D) circCHRNG wild-type (WT) or mutant (MUT) luciferase reporters and miR-133 mimics, or mimic negative control (NC), were co-transfected in 293T cells. (E) Expression of miR-133 in ovine SMSCs transfected with si-NC and si-circCHRNG. (F) Protein expression of SRF and MEF2A in ovine SMSCs transfected with si-NC and si-circCHRNG. (G) Expression of PAX7 protein in ovine SMSCs infected with miR-NC miR-133 mimics and miR-133 inhibitor. (H) MTT assays for ovine SMSCs co-transfected with si-circCHRNG and miR-133 inhibitor or miR-NC. (I) Expression of PAX7 protein in ovine SMSCs co-transfected with si-circCHRNG and miR-133 inhibitor or miR-NC. (J) Expression of SRF and MEF2A protein in sheep SMSCs co-transfected with miR-133 inhibitor and si-circCHRNG or si-NC. We considered p < 0.05 to be statistically significant. * p < 0.05, ** p < 0.01.