Abstract
As our “hidden organ”, microbes widely co-exist at various sites on the human body. These microbes are collectively referred to as the microbiome. A considerable number of studies have already proven that the microbiome has significant impacts on human health and disease progression, including cancers. The recent discovery of cancer-specific microbiomes renders these cancer-associated microbes as potential biomarkers and therapeutic targets. While at low biomass levels, the lung microbiome still dramatically influences the initiation, progression and treatment of lung cancers. However, research on lung cancer-associated microbiomes is emerging, and most profiling studies are performed within three years. Unfortunately, there are substantial inconsistencies across these studies. Variations in microbial diversity were observed, and different microbial biomarkers for lung cancer have been proposed. In this review, we summarized the current findings of lung cancer microbiome studies and attempt to explain the potential reasons for the dissimilarities. Other than lung microbiomes, oral and airway microbiomes are highly related to lung microbiomes and are therefore included as well. In addition, several lung cancer-associated bacterial genera have been detected by different independent studies. These bacterial genera may not be perfect biomarkers, but they still serve as promising risk factors for lung cancers and show great prognostic value.
Keywords: lung cancer, microbiome, biomarker, respiratory tract, lung dysbiosis
1. Introduction
A vast number of microorganisms, or microbes, colonize various human body sites, such as the gut, skin, mouth and respiratory tract. Such microorganisms include (but are not limited to) bacteria, yeasts, fungi and viruses, and they are collectively known as the microbiome or microbiota. While microbiota refer to microorganisms in a specific environment, the microbiome describes a collection of genomes from all microorganisms in the environment. As this review mainly discusses the genomic profiling of lung cancer-associated microorganisms, the term “microbiome” is used. These microbes are at least as abundant as the somatic cells in an individual [1] and contain far more genetic information than a human genome [2]. Numerous studies have revealed the essential roles of the microbiome in health and disease via the modulation of signaling transduction, metabolism, tissue homeostasis and immune responses [3,4,5,6,7].
It is not surprising that the microbiome influences cancer progression and treatment [8]. For example, Helicobacter pylori is a well-known carcinogen contributing to gastric cancer initiation [9]. Fusobacterium nucleatum is also closely related to the invasion and metastasis of colorectal cancer [10]. Recently, Nejmen et al. [11] performed a comprehensive analysis of 1526 tumors and paired paracancerous normal tissues across seven tumor types and discovered that different tumor types harbor distinct intratumor microbiome compositions. These intratumor bacteria correlate with the patient’s underlying diseases, status and response to treatment, suggesting the use of tumor-resident microbiomes as diagnostic biomarkers. Moreover, a recent finding suggests that even low abundant intratumor microbes can reshape the cytoskeleton and promote the survival of circulating tumor cells, leading to tumor metastasis [12]. This indicates that the tumor-resident microbiome is not only a result of tumor formation, but they also actively contribute to cancer development. Furthermore, recurrent HLA-I-bound peptides derived from tumor-residing intracellular bacteria were identified on tumor cells in melanoma patients, and they can induce the activation of tumor-infiltrating T cells [13]. Such findings strongly support targeting specific intratumor bacteria in anti-tumor immunotherapy.
Lung cancer is currently the global leading cause of cancer-caused death, claiming 1.80 million lives in 2020 [14]. Given the crucial role of the cancer-resident microbiome in cancer progression and diagnosis, it would be worth investigating the microbiome of lung cancer tissues as well. Such microbiomes can be potential microbial biomarkers of lung cancers and may contribute to tumorigenesis. However, lung tissues are difficult to acquire and it is almost impossible to obtain the control tissues from healthy individuals. Therefore, due to their close relationship (which will be elaborated upon later), many studies also used samples from the mouth or other respiratory tract sites to study the lung cancer-associated microbiome. In this review, we focus on lung cancer-associated microbiome profilings in the mouth and respiratory tract (including the lung). The potential bacterial biomarkers are introduced, and the reasons for dissimilarities across studies are discussed.
2. Lung or Tumor-Resident Microbiome and Lung Cancers
While human lungs are constantly exposed to the environment and microbes, healthy lungs were traditionally considered sterile organs (i.e., free of resident bacteria) [15]. However, since the development of culture-independent molecular techniques such as 16s rRNA sequencing, numerous studies revealed many lung-residing microorganisms (i.e., bacteria, viruses, and fungi). In healthy lungs, such lung microbiomes are in low abundance, with a much lower microbial biomass than other body sites [16]. Despite the low microbial burden, the lung microbiome is still paramount for body homeostasis and health, including the modulation of inflammation [17,18] and pathogen or pollutant protection [11,19].
As for lung cancers, the newly emerged spatial meta-transcriptomics revealed that the bacterial load in lung cancer cells is significantly higher than that in the surrounding stromal cells and immune cells [20]. Moreover, a clear decreasing trend in bacterial load was observed from the airways to lung cancer cells to tertiary lymphoid structures and the adjacent normal lung tissues [20] (or upper airways to lower airways and lung [21]). Similarly, oral taxa were identified in bronchoscopy and lobectomy samples, with a higher abundance in bronchoscopy samples [22]. Therefore, lung cancer-resident bacteria likely originate from environmental exposure and the oral/airway microbiome rather than the colonization of gut-derived bacteria from blood (which is another primary source of intratumor bacteria for many cancer types). It should be noted that there are substantial inconsistencies across studies of lung cancer microbiomes (which will be elaborated in Section 2.1 and Section 2.2); the findings concerning bacterial load in lung cancer should be evaluated with caution. Nevertheless, the lung-resident microbiome has great diagnostic and prognostic potential, and the profiling of the lung cancer-resident microbiome is reviewed herein (Table 1).
2.1. Few Commonalities across Different Lung Microbiome Profilings
In one of the earliest characterizations of human lung tissue and lung cancer microbiome using a large cohort, Yu et al. [23] analyzed non-malignant lung tissue samples from 165 lung cancer patients and tumor samples from 31 patients. They identified unique microbial communities in the lung, with the dominant phylum Proteobacteria. Moreover, the abundance of genus Thermus and Legionella in paracancerous lung tissues is elevated in patients with advanced stages and metastasis. Similarly, the abundances of Thermus and Ralstonia differ in different lung cancer subtypes (i.e., lung adenocarcinomas and lung squamous cell carcinoma). Therefore, these bacteria may serve as potential biomarkers for lung cancer prognosis. However, numerous studies following lung cancer-associated microbiome profilings do not provide similar findings. Such dissimilarities occur in the specific tumor-resident bacteria identified and involve the overall microbiome diversity or richness.
Table 1.
Summary of studies of lung cancer-associated microbiome in the oral cavity and respiratory tract (most recent first).
| Study | Sampling Site | Disease | Experiment Design | Sequencing Methods * | Diversity Variations in LC | Microbial Associations or Biomarkers |
|---|---|---|---|---|---|---|
| Yuan [24] | Tumor | LC | RM LC (n = 174) vs. non-RM LC (n = 134) | TCGA | RM LC has similar α-diversity, but reduced richness | Acidovorax, Clostridioides, Succinimonas, Shewanella, Leuconostoc and Dickeya are biomarkers for RM LC |
| Baranova [25] | Sputum | LUSC | Patients (n = 40) vs. healthy controls (n = 40); all male | 16S V3–V4 | Decreased β-diversity in LUSC; no changes in α-diversity | Streptococcus, Bacillus, Gemella and Haemophilus are enriched in LUSC patients |
| Wu [26] | BALF; tumor | LC as GGO | BALF from diseased lung and paired contralateral healthy lung (n = 11); lung GGO and paired adjacent normal tissues (n = 26) | 16S V4/16S V3/16S V3–V4/16S V4–V5 | No changes in α- and β-diversity | Significantly reduced Proteobacteria in LC tissues; In BALF of LC patients: reduced Rothia, and increased Lachnospiraceae, Bacteroides uniforms and Faecalibacterium prausnitzii |
| He [27] | Sputum | NSCLC and/or COPD | patients with NSCLC and COPD (CN, n = 67) vs. NSCLC (n = 9) vs. COPD (n = 14) | 16S V3–V4 | No significant differences in diversities | In CN patients: reduced Streptococcus, Veillonella, Moraxella and Actinomyces; and increased Neisseria and Acinetobacter |
| Vogtmann [28] | Oral wash | LC | Patients (n = 1306) | 16S V4 | Higher α-diversity is associated with lower LC risk | Increased Streptococcus and Peptoniphilus abundances are associated with increased LC risk, while Peptostreptococcus, Eubacterium yurii and Aggregatibacter are associated with reduced risk |
| Kim [29] | Tumor | NSCLC | Tumor tissue (n = 162) vs. adjacent normal tissue (n = 54) | 16S V4–V5 | Reduced diversity as LC progress | Increased Romboutsia, Novosphingobium, Acinetobacter and Prevotella in LC Increased Stenotrophomonas upon LC relapse |
| Qian [30] | BALF; airway protected brushing | sMPLC | Patients (n = 8) | 16S V3–V4 | Increased α-diversity in BALF | Clostridium, Actinobacteria, Fusobacterium and Rothia are enriched in the BALF of sMPLC lesions |
| Chen [19] | Tumor | LC | Tumor tissue (n = 34) vs. adjacent normal tissue (n = 29) | 16S | Lower α-diversity and higher β-diversity in LC tissues | In LC: increased Staphylococcus, Capnocytophaga, Lachnoanaerobaculum, Fusobacterium, Oligella, Rubellimicrobium, Marinococcus Sphingomonas and Sphingopyxis; and decreased Comamonas and Peptococcus |
| Masuhiro [31] | BALF | LC | Patients under PD-1 blockade treatment (n = 12) | 16S V3–V4 | Higher diversity in responders | Responders have higher Bacteroidetes and lower Proteobacteria |
| Zhang [32] | BALF; tumor | NSCLC | Patients (n = 6 for BALF; n = 37 for tumor tissues) | Pathogen-targeted sequencing (tumor and 4 BALF); 16S (2 BALF) | Higher diversity in BALF than in tumor | BALF and tumor tissues share Streptococcus pneumoniae, S. crista, S. constellatus, S. gordonii, Prevotella II, Haemophilus, H. haemolyticus, H. influenzae, Actinomyces Neesii, human herpes virus type 7 and Neisseria lactose |
| Marshall [33] | Epithelial brushing | Pre-cancer | A 10-year follow-up study of 393 patients: with incidence (n = 59), prevalence (n = 21), and no cancer (313) | 16S V4 | NA | The abundances of Bacilli, Lactobacillales, Streptococcus and Paenibacillus are associated with incident LC |
| Zeng [34] | BALF | NSCLC | LC (n = 46) vs. benign lung disease (n = 29) | 16S V3–V4 | Increased α-diversity during carcinogenesis and significant changes in β-diversity | Enrichment of phyla (Firmicutes and Bacteroidetes) and genera (Streptococcus, Prevotella and Veillonella) in NSCLC |
| Chu [35] | BALF | LC | Patients under PD-1 blockade treatment: responders (n = 19) vs. non-responders (n = 27) | 16S V3–V4 | Decreased diversity upon treatment | Increased abundance of Fusobacterium is associated with poor anti-PD-1 therapy response |
| Zhang [36] | Tumor | NSCLC | Patients (n = 53) | Pathogen-targeted sequencing | NA | At advanced stage: increased Serratia marcescens, Actinomyces neesii, Enterobacter cloacae and Haemophilus parainfluenzae; and decreased Staphylococcus haemolyticus and Streptococcus crista Survival prediction: Haemophilus parainfluenzae, Serratia marcescens, Acinetobacter jungii and Streptococcus constellation High PD-L1 expression: increased Acinetobacter jungii |
| Huang [37] | Sputum | NSCLC | Patients (n = 85) | 16S V3–V4 | Decreased α- and β-diversity at advanced stage | Early stage: Granulicatella and Actinobacillus are enriched Advanced stage: Actinomyces is enriched |
| Roy [38] | Saliva | LUAD | Patients (n = 5) and healthy control (n = 5) | 16S V3–V4 | No significant changes in α-diversity | Increased Rothia mucilaginosa, Veillonella dispar, Prevotella melaninogenica, Prevotella pallens, Prevotella copri, Haemophilus parainfluenzae, Neisseria bacilliformis and Aggregatibacter segnis in LUAD |
| Dong [39] | Tumor | LC | Tumor tissues (n = 118) vs. adjacent normal tissue (n = 123) from 143 patients | 16S V3–V4 | No significant changes in α-diversity but significant differences in β-diversity |
Massilia, Phenylobacterium and Pseudoxanthomonas are enriched in tumor tissue; Brevibacillus, Cupriavidus and Anaerococcus are enriched in normal tissues Massilia and Acidovorax are associated with TP53 mutation |
| Jang [40] | BALF | LC | Patients under PD-1 blockade treatment (n = 84) | 16S V3–V4 | No significant changes in α-diversity and β-diversity | High-PD-L1 group: dominated by Veillonella dispar; with reduced Neisseria Responders: dominated by Veillonella dispar Non-responders: dominated by Haemophilus influenzae and Neisseria perflava |
| Boesch [41] | Tumor | AdvancedNSCLC | Tumor tissues (n = 38) vs. adjacent normal tissue (n = 10) from patients with PD-1 blockade treatment | 16S V3–V4 | Increased diversity is associated with better survival | Gammaproteobacteria correlate with low PD-L1 expression and poor anti-PD-1 blockade treatment outcomes |
| Lu [42] | Sputum | NSCLC | Patients (n = 87) vs. healthy controls (n = 34) | 16S V3–V4 | Decreased α-diversity in NSCLC | NSCLC: increased Haemophilus parainfluenzae and Haemophilus influenzae Distant metastasis: decreased Capnocytophaga; and increased Pseudomonas, Coriobacteriaceae and Actinomyces |
| Chang [43] | Tumor | LC | Patients (n = 49) | 16S V4 | NA | Brevundimonas diminuta, Acinetobacter radioresistens Enterobacter cloacae, Mycobacterium chelonae, Mycobacterium franklinii, Staphylococcus sp., Bacillus megaterium, Pseudomonas aeruginosa and Rhodococcus erythropolis are enriched in LC and associated with poor prognosis |
| Shi [44] | Mouth rinse | LC | Patients (n = 156) vs. healthy control (n = 156) | 16S V4 | No significant changes in α-diversity and β-diversity | The abundances of families Lachnospiraceae, Peptostreptococcaceae, Erysipelotrichaceae and species Parvimonas micra are associated with decreased LC risk |
| Seixas [45] | BALF | LC, COPD and ILD | LC (n = 8) vs. COPD (n = 7) vs. ILD (n = 10) | 16S V4 | No significant changes in diversity between cancer and non-cancer |
Streptococcus and Prevotella are associated with LC Haemophilus is associated with COPD |
| Zheng [22] | BALF | NSCLC | Patients (n = 32) vs. non-cancer controls (n = 15) | 16S | Decreased diversity in NSCLC | LC: increased Lactobacillus rossiae, Burkholderia mallei and Bacteroides pyogenes; decreased Paenibacillus odorifer, Pseudomonas entomophila and Magnetospirillum gryphiswaldense |
| Zhang [46] | Sputum; stool | Metastatic NSCLC | Patients (n = 75) at baseline and during immune checkpoint inhibitors treatment | 16S | α-diversity between the gut and respiratory microbiota is not related Only increased α-diversity in the gut is associated with better treatment outcomes |
Streptococcus in sputum as a biomarker for good treatment response |
| Dumont-Leblond [47] | Tumor | NSCLC | Tumor tissues vs. adjacent normal tissue from 29 patients | 16S V3–V4 | Higher β diversity differences among different patients than tissues from the same patient. Higher α-diversity in tumor tissues |
LC has an increased abundance of pathogenic and pro-inflammatory bacteria: Escherichia-Shigella, Faecalibacterium, Pseudomonas, unclassified Enterobacteriaceae, Alloprevotella and Brevundimonas High Phascolarctobacterium in LUSC |
| Ma [48] | Tumor | LUAD as SSN or SN | Tumor tissues vs. adjacent normal tissue (n = 10 pairs for SSN; n = 25 pairs for SN) | 16S V3–V4 | SSN has higher microbiome richness and diversity Tumor and normal tissues have similar diversity and richness |
Increased Actinobacteria, Proteobacteria, Parvibaculales, Parvibaculaceae, Parvibaculum, Renibacterium and Ancylobacter; and decreased Firmicutes, Bacteroidetes and Lactobacillus in LUAD |
| Leng [49] | Tumor; sputum | LC | Tumor tissues vs. adjacent normal tissue (n = 31 pairs); sputum from NSCLC patients (n = 17) vs. cancer-free smoker controls (n = 10) | Droplet digital PCR for 25 NSCLC-associated bacterial genera | NA | Enrichment of Acidovorax, Streptococus and Veillonella in sputum of LUSC Enrichment of Capnocytophaga in sputum of LUAD |
| Druzhinin [50] | Sputum | LC | Patients (n = 66) vs. healthy controls (n = 62); all male | 16S V3–V4 | Decreased β diversity in LC patients | Increased Streptococcus, Bacillus, Gemella and Haemophilus in LC patients Chromosomal aberration frequency is positively associated with increased Bacteroides, Lachnoanaerobaculum, Porphyromonas, Mycoplasma and Fusobacterium; and decreased Granulicatella. Micronuclei frequency is negatively associated with increased Megasphaera and Selenomonas bovis |
| Hosgood [51] | Oral rinse | LC | Patients (n = 114) vs. healthy controls (n = 114) | Metagenomic shotgun sequencing | Individuals with lower α-diversity had an increased risk of lung cancer No significant changes in β-diversity |
Decreased risk of LC: a higher abundance of Spirochaetia and Bacteroidetes Increased risk of LC: Bacilli class and Lactobacillales order |
| Tsay [52] | Lower airway brushing; buccal brushing | LC | Patients (n = 83) | 16S V4 | α-diversity is similar across different stages of NSCLC. Higher α-diversity in lower airways than in upper airways |
Veillonella parvula is associated with LC progression, IL-17 expression and the activation of the immune checkpoint Increased Moraxella, Fusobacterium, Pseudomonas and Haemophilus; and decreased Actinomycetales in advanced LC Streptococcus, Prevotella and Veillonella enrichment is related to poor prognosis |
| Zhuo [53] | BALF | LC | From cancerous lung and the contralateral non-cancerous lung (n = 50) | 16S V3–V4 | No significant changes in α- and β-diversity | Increased risk of LC: genera Weissella and Spiroplasma Decreased risk of LC: phylum Bacteroidetes (class Bacteroidia and order Bacteroidales) |
| Kovaleva [54] | Tumor | NSCLC | Tumor tissues vs. adjacent normal tissue (n = 89) | 16S V3–V4 | Tumor tissues have similar α-diversity, but reduce overall bacterial load | High bacterial load with increased iNOS expression is a favorable prognostic factor; High bacterial load with increased FOXP3+ cells is associated with poor prognosis Increased Propionibacterium is associated with lower iNOS expression |
| Cheng [55] | BALF | LC | Patients (n = 32) vs. benign pulmonary diseases (n = 22) | 16S V3–V4 | Similar richness and evenness in LC | TM7-3, Gemmiger, Capnocytophaga, Sediminibacterium, Blautia and Oscillospira are enriched in LC |
| Mao [56] | Tumor | LC | Tumor tissues vs. adjacent normal tissue (n = 55) | 16S V3–V4 | Reduced α-diversity in LC; but no significant changes in β-diversity |
Propionibacterium is significantly reduced in tumor tissues Other reduced genera include: unclassified Comamonadaceae, unclassified Enterobacteriaceae, Rhodobacter, Psychrobacter, Phormidium, Propionibacterium, Microbacterium and Finegoldia |
| Bello [57] | Bronchial biopsy; saliva | Central LC | Patients (n = 25): saliva and biopsies of affected and contralateral bronchi vs. healthy controls (n = 16): saliva and single bronchi biopsy |
16S V3–V4 | The diversity of salivary sample is comparable in patients and controls |
Streptococcus has dominated in both affected and contralateral bronchi of patients Pseudomonas is dominated in control Increased abundance of Streptococcus, Rothia, Gemella and Lactobacillus in patients’ saliva |
| Druzhinin [58] | Sputum | LC | Patients (n = 17) vs. healthy control (n = 17) | 16S V3–V4 | No significant differences in α-diversity | Increased genera Haemophilus and Bergeyella; and decreased genera Atopobium, Stomatobaculum, Treponema and Porphyromonas in LC patients Chromosomal aberration frequency is negatively associated with the genus Atopobium and positively associated with the species Alloprevotella |
| Wong [59] | Tumor | LC | Tumor tissues vs. adjacent normal tissue (n = 497 for LUAD and 433 for LUSC) | TCGA | NA | The LC-associated microbiome is age and gender-specific Escherichia coli str. K-12 substrain W3110 is associated with the survival of aged LUAD patients |
| Reinhold [21] | Tumor; PO swab; BALF | LC | Patients undergoing surgery (n = 13) | 16S V3–V4 | Decreased α-diversity in the upper airways | High Prevotella, Veillonella and Streptococcus in the upper airways and BALF High Pseudomonas, Propionibacteria, Proteobacteria and Actinobacteria in lung cancer tissues |
| Bingula [60] | Saliva; BAL (from excised lobe); tumor | NSCLC | saliva, BAL, peritumoral tissues, tumor tissues and adjacent normal tissue from 18 patients | 16S V3–V4 | Unique β-diversity of BAL Diversity varies depending on lobe location |
Tissue samples: dominated by Phylum Proteobacteria
BAL: dominated by class Clostridia Saliva: dominated by class Bacilli |
| Patnaik [61] | Saliva; BALF; tumor | Early recurrentNSCLC | Pre-surgery saliva and BALF; tumor tissues and adjacent normal tissue from 48 patients undergoing surgery | 16S | Higher diversity in saliva and BALF; Tumor tissues and adjacent normal tissue have similar diversity |
Recurrence is associated with increased genus Delftia and decreased Bifidobacterium in saliva; as well as increased Staphylococcus and decreased Bacillus and Anaerobacillus in tumor tissues |
| Ekanayake [62] | BALF; PO swab | LC and BRS | Patients (n = 20 for LC and n = 20 for BRS) vs. healthy controls (n = 20) | 16S V3–V4 | Increased diversity in patients | Enterococcus faecalis, Corynebacterium tuberculostearicum and Keratinibaculum paraultunense are LC-specific |
| Huang [63] | Bronchial washing fluid; sputum | LC | Bronchial washing fluid (n = 40) and sputum (n = 52) from LC patients | 16S V3–V4 | No significant difference in α- and β- diversity between LUAD and LUSC | All from Bronchial washing fluid samples: Genera Veillonell, Megasphaera, Actinomyces and Arthrobacter are enriched in LUAD without metastasis Genera Capnocytophaga and Rothia are enriched in LUSC with metastasis Streptococcus is decreased in LUAD upon metastasis Veillonella and Rothia are increased in LUSC upon metastasis |
| Jin [64] | BALF | LC | Patients (n = 91) vs. nonmalignant pulmonary diseases (n = 29) vs. healthy controls (n = 30); a validation cohort of 85 patients |
Metagenomics | Diversity and richness are reduced in LC patients β-diversity is different between LC patients and healthy controls |
Haemophilus influenzae shows the greatest difference between LC patients and healthy controls |
| Gomes [65] | BALF | LC | Patients (n = 49) vs. healthy controls (n = 54) | 16S V3-V6 | LUSC has a higher diversity than LUAD | Biomarkers for LUAD: Acinetobacter, Propionibacterium, Phenylobacterium, Brevundimonas and Staphylococcus Biomarkers for LUSC: Enterobacter, Serratia, Klebsiella, Kluyvera, Morganella, Achromobacter and Capnocytophaga |
| Ren [66] | Tumor | LUAD as GGN | Tumor tissues (n = 10) vs. adjacent normal tissue (n = 5) | Whole genome sequencing | High β diversity variation among patients | No significant differences in microbiome compositions between GGNs and normal tissues (except LUAD) |
| Zhang [67] | Saliva | NSCLC | Patients (n = 39) vs. healthy controls (n = 20) | 16S V1-V2 | A higher richness and lower diversity in NSCLC patients | In NSCLC: increased Veillonella, Streptococcus, Lautropia, Leptotrichia, Rothia and Aggregatibacter; and decreased Prevotella_7, Fusobacterium, Porphyromonas, Alloprevotella, Prevotella, Bacteroides and Faecalibacterium Veillonella is positively associated with the Neutrophil-lymphocyte ratio Streptococcus is negatively associated with the lymphocyte-monocyte ratio |
| Wang [68] | Saliva; BALF | PBC | Patients (n = 51) vs. healthy controls (n = 15) | 16S V4 | Patients have lower diversity in both saliva and BALF samples | Treponema (in saliva) and Filifactor (in both saliva and BALF) are potential biomarkers for LC |
| Hosgood [69] | Sputum | LC | Patients (n = 45) vs. healthy controls (n = 45) | 16S V1-V2 | Lower α-diversity is associated with an increased risk of LC | Decreased relative abundance of Fusobacteria is a risk factor for LC |
| Peters [70] | Tumor | NSCLC | Tumor tissues vs. remote normal tissue (n = 19 pairs) | 16S V4 | Tumor tissues have reduced richness and diversity | Increased Koribacteraceae; and decreased Bacteroidaceae, Lachnospiraceae and Ruminococcaceae in normal tissues are associated with a better survival outcome |
| Yang [71] | Saliva | LC | Patients (n = 75) vs. healthy controls (n = 172); all female | 16S V1-V2 | Tumor tissues have reduced richness and diversity | Increased Sphingomonas and Blastomonas in LC patients |
| Liu [72] | Tumor | LC | LC-only (n = 11) vs. emphysema-only (n = 10) vs. both LC and emphysema (n = 19); all heavy smokers | 16S V4 | The emphysema-only group has a lower diversity | LC vs. emphysema-only: decreased Proteobacteria (primarily the genera Acinetobacter and Acidovorax); and increased Firmicutes (Streptococcus) and Bacteroidetes (Prevotella) |
| Greathouse [73] | Tumor | LC | Patients (n = 143) vs. healthy controls (n = 33) TCGA was used as a validation cohort |
16S V3-V5 | Control tissues have lower α-diversity | Acidovorax, Klebsiella, Rhodoferax and Anaerococcus are enriched in LUSC only |
| Tsay [74] | Lower airway brushing; buccal brushing | LC | Patients (n = 39) vs. non-cancer patients (n = 36) vs. healthy controls (n = 10) | 16S V4 | No differences (α- and β-diversity) in buccal samples Significant changes in β-diversity in Lower airway samples between LC and non-cancer/healthy controls |
Streptococcus and Veillonella are highly enriched in the lower airways of LC patients and are associated with ERK and PI3K signaling pathway activation |
| Liu [75] | Bronchial specimen brushing | LC | Diseased lung and paired contralateral healthy lung (n = 24 pairs) vs. healthy controls (n = 8) | 16S V3–V4 | α-diversity reduces from healthy site to noncancerous to cancerous site | Genera Streptococcus and Neisseria are significantly more abundant in LC Genera Staphylococcus and Dialister are significantly more abundant in healthy controls |
| Cameron [76] | Sputum | LC | Patients (n = 4) vs. non-cancer controls (n = 6) | 16S | No significant changes in α-diversity | Streptococcus viridans and Granulicatella adiacens are significantly increased in LC patients |
| Lee [77] | BALF | LC | Patients (n = 20) vs. benign diseases (n = 8) | 16S V1-V3 | Increased diversity in LC |
Phyla Firmicutes and TM7 are significantly increased in LC patients |
| Yu [23] | Tumor | LC | Tumor tissues (n = 31) vs. remote normal tissue (n = 165) | 16S V3–V4 | α-diversity is increased with environmental exposures, residence population, smoking and disease history LC has reduced diversity |
Biomarkers for advanced LC: Genus Thermus Biomarkers for LC metastasis: Genus Legionella |
| Yan [78] | Saliva | LC | Patients (n = 10 for LUAD and n = 10 for LUSC) vs. healthy controls (n = 10) | 16S V3 and V6 | NA |
Capnocytophaga and Veillonella are promising biomarkers for LUSC The abundance of Capnocytophaga, Selenomonas, Veillonella and Neisseria in saliva is significantly changed in LC patients |
| Hosgood [79] | Sputum; oral rinse | LC | Patients (n = 8) vs. healthy controls (n = 8) | 16S V1-V2 | The diversity between LC and control is similar in buccal samples, but significantly different in sputum | Granulicatella, Abiotrophia and Streptococcus are enriched in the sputum of LC patients |
To have a complete collection of lung cancer-associated microbiome studies, the search term in PubMed is (“lung cancer” [Title/Abstract] OR “lung neoplasm” [Title/Abstract] OR “pulmonary neoplasm” [Title/Abstract] OR “pulmonary cancer” [Title/Abstract] OR “lung adenocarcinoma” [Title/Abstract] OR “pulmonary adenocarcinoma” [Title/Abstract] OR “lung squamous cell carcinoma” [Title/Abstract] OR “squamous cell lung carcinoma” [Title/Abstract] OR “lung large cell carcinoma” [Title/Abstract] OR “large cell lung carcinoma” [Title/Abstract] OR “lung small cell carcinoma” [Title/Abstract] OR “small cell lung carcinoma” [Title/Abstract]) AND (“microbiome” [Title/Abstract] OR “Metagenome” [Title/Abstract] OR “microbiota” [Title/Abstract] OR “microbe” [Title/Abstract]). Meta-analysis, reviews and studies about gut microbiome only are excluded. Only profilings of the microbiome in the oral cavity and respiratory tract were selected, and 60 studies were included (as of 2022, Oct 14). LC: lung cancer; RM: recurrent/metastasis; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; NSCLC: non-small cell lung cancer; COPD: chronic obstructive pulmonary disease; ILD: interstitial lung disease; BAL: bronchoalveolar lavage; BALF: bronchoalveolar lavage fluid; GGO: ground-glass opacity; sMPLC: synchronous multiple primary lung cancer; SSN or SN: subsolid nodules or solid nodules; PO: posterior oropharynx; BRS: bronchiectasis; PBC: primary bronchogenic carcinoma; GGN: ground-glass nodules. * For 16S sequencing, some studies failed to provide the hypervariable regions they sequenced.
Microbiome richness and diversity are two important parameters for characterizing microbial communities as microorganisms behave and function as communities. Therefore, two microbiome diversities, α-diversity and β-diversity, are often assessed. The α-diversity is the structure of a microbial community concerning its richness (i.e., number of taxa) and/or evenness (i.e., distributions of abundances of these taxa). On the contrary, β-diversity compares the microbiome’s composition between samples. In most cases, a healthy microbiome has higher α-diversity. Indeed, many studies reported decreased diversity or richness in the lung cancer microbiome compared to controls (Table 1). For example, in a study comprising 162 non-small cell lung cancer (NSCLC) patients, the α-diversity of NSCLC tissues was significantly lower than its adjacent normal lung tissues. Moreover, the α-diversity keeps decreasing as the disease progresses [29]. Similarly, although recurrent or metastatic lung cancer has a similar α-diversity as non-recurrent or metastatic lung cancer, its microbial richness is significantly reduced [24]. Regarding anti-PD-1 therapy, reduced lung cancer microbiome diversity is also related to worse clinical outcomes and poorer patient survival [41]. On the contrary, an almost equal number of studies suggested that lung cancers have a similar (or increased) microbial diversity compared to various controls (e.g., tissues from healthy volunteers, adjacent paracancerous normal tissues or normal tissues distant from tumors). For instance, Dong et al. [39] analyzed lung cancer tissues and normal control tissues that were distant from the tumors of a large cohort of 143 patients. They reported that the α-diversity of lung cancers and distant normal lung tissues is not significantly different (although there are significant differences in β-diversity between both groups). Similarly, in two independent studies with two distinct cohorts of 89 non-small cell lung cancer (NSCLC) patients [54] and 35 lung adenocarcinomas (LUAD) [48], the microbiomes of lung cancer tissues and their adjacent normal tissues exhibit similar α-diversity as well. Therefore, there are substantial inconsistencies across these studies, significantly increasing the difficulty in identifying lung cancer-specific biomarkers.
2.2. Potential Reasons for the Few Commonalities across Studies
Such a lack of consensus across studies may be related to multiple factors, including sampling techniques, sequencing methods, the choice of controls, and most importantly, environmental and host variations. Firstly, as aforementioned, the low microbial biomass of the lung renders it very sensitive to contamination when handling the sample. However, in most studies, the microbiome of lung cancer tissues is compared to that of the adjacent paracancerous “normal” lung tissues. These samples experience similar background contamination during processing; therefore, unique lung cancer-resident bacteria can still be identified. This suggests that potential contamination may not be the primary cause of the few commonalities across studies.
Secondly, although most studies used16S rRNA gene sequencing for bacteria identification, different variable regions of 16S rRNA were sequenced in these studies (Table 1). There are nine hypervariable regions (V1–V9) in bacterial 16S rRNA genes, and V1, V2, V3, V4, and/or V5 were mostly sequenced for detecting lung microbiomes. However, no single region can differentiate all bacteria, and each region has its relative advantages in identifying different bacterial species [80]. It would be ideal to sequence multiple regions of 16S rRNA genes, as Wu et al. [26] performed in their study. Other than 16S rRNA sequencing, targeted pathogen sequencing [32,36,49,81], whole-genome sequencing (WGS) [66] or RNA-seq (which may be obtained from TCGA database) [24,73,82,83] were used to identify lung cancer microbiomes. The application of targeted pathogen sequencing may miss some novel bacteria species, and the ability to detect microbes using RNA-seq or WGS dramatically depends on the sequencing the depth of the experiments. Therefore, it is inevitable that dissimilarities exist across studies.
Thirdly, various control samples were used for comparison in these studies. As introduced earlier, tissues from healthy volunteers or benign lung diseases, adjacent paracancerous normal tissues, normal tissues distant from tumors or contralateral noncancerous tissues were used as baseline references (Table 1). However, the microbiome’s diversity varies among these controls, and the previous review indicated that this might be the primary cause of variations among the studies [84]. Indeed, Greathouse et al. [73] reported that the α-diversity is lower in normal lungs compared to adjacent paracancerous normal tissues. However, in another study with a similar design, Liu et al. [75] observed that the α-diversity decreased from healthy controls to the contralateral noncancerous lung of lung cancer patients to lung cancer tissues. Moreover, contradictory findings were provided for studies using adjacent paracancerous normal tissues as controls (refer to the examples provided previously [29,48,54]). These findings indicate that the choice of the control is not the underlying reason for the lack of consensus observed in lung microbiome profiling.
Fourthly and most importantly, as the lung is under constant environmental exposure, environmental factors would be the primary cause of variations in the lung microbiome. This could also explain the inconsistent results observed in various controls. Indeed, in their analysis of the impact of smoking and indoor air pollution (i.e., coal burning) on the lung cancer microbiome, Chen et al. [19] found that the pollutants significantly decreased the biodiversity of lung tissue-specific microbes, especially in paracancerous normal tissues. Pollutant-detoxication microbes (such as Sphingomonas and Sphingopyxis) are highly enriched in the lungs to protect their lungs under such conditions. Similarly, Dong et al. [39] reported that polycyclic aromatic hydrocarbon (PAH)-degrading microbes, Massilia and Sphingobacterium, are more abundant in lung cancer tissues of smokers than non-smokers. Hosgood et al. [79] reported similar findings on coal-burning pollutions as well. Interestingly, the abundance of another PAH-degrading microbe, Acidovorax, is cancer subtype-specific. It is only highly enriched in the lung squamous cell carcinoma (LUSC) tissues (and not LUAD tissues) of smokers [39,73]. In a large cohort analysis of lung microbiome in 165 noncancerous lung tissues from lung cancer patients, the α-diversity of the lung microbiome is dependent on various environmental factors: air pollutions/air particulate matters, the population density of residential area, and smoking or lung disease histories [23]. Notably, even tobacco smoking and electronic cigarette usage also result in different microbial communities in the lungs [85]. These findings indicate that the lung microbiome is sensitive to even very subtle environmental changes. Researchers should consider all these identified and unidentified environmental factors when analyzing lung microbiome data.
Finally, studies have suggested high β-diversity differences in lung cancer microbiome among patients [66]. Host or patient variations are also significant contributors to the dissimilarities of different lung microbiome profilings, although some host variations are closely related to environmental factors. As aforementioned, LUSC and LUAD have different enriched specific tumor-resident bacteria (e.g., Acidovorax) [39,73]. This is further confirmed by another analysis of 497 LUAD and 433 LUSC patients, which indicated that both LUAD and LUSC tumor tissues contain unique microbiomes compared to their adjacent paracancerous tumor tissues [59]. Moreover, the authors reported that such lung cancer microbiomes are also age- and gender-specific. In older LUAD patients (both male and female), Escherichia coli strain K-12 substrain W3110 dysregulation is a promising prognostic marker for patient survival. On the contrary, Pseudomonas putida strain KT2440 is uniquely enriched in young LUSC male patients. Similarly, Chen et al. [19] also reported that females have a higher α-diversity than males in both tumor tissues and adjacent paracancerous normal tissues. Other than tumor subtypes, gender and age, gene/pathway mutation-specific microbes are also identified. For example, lung cancers with EGFR mutation are associated with the absence of Pseudomonas. aeruginosa [43], the increased abundance of Serratia marcescens or the decreased abundance of Haemophilus parainfluenzae [36]. However, it remains to be determined whether such gene/pathway mutations lead to specific microbe enrichments or whether specific microbes drive certain gene mutations. Nevertheless, these specific microbes can still be used as potential prognostic biomarkers.
In conclusion, the lung microbiome is very sensitive to external variations. Studies have already begun to identify lung cancer-associated microbes in patients with specific conditions (e.g., heavy smokers [72], never smokers [51] or females only [71]). However, larger cohorts with much more complex patient stratifications are still required to thoroughly examine the association of specific microbes with lung cancers in patients under various conditions.
3. Airway or Respiratory Tract Microbiome and Lung Cancers
As lung biopsy is hard to access and obtaining “true health” lung tissues in most cases is unethical, numerous studies with respect to lung microbiomes are based on bronchoalveolar lavage, bronchial washing, sputum or oral samples. There were concerns about the potential contamination of the lung microbiome from airway/oral bacteria [86]. However, as introduced earlier, the lung microbiome is closely related to (and maybe even originated from) the airway’s microbiome [20,22], which also reflects the spatial relationship of different sampling sites [61]. Therefore, the airway’s microbiome can still represent lung-resident microbial communities. While variations (even significant variations) exist among microbiomes at different sites of airways/mouth [21,22,26,60,63,73,81], studies still identified lung cancer-specific bacteria using these samples (Table 1). Unfortunately, a lack of consensus still exists among these studies on airway microbiomes due to similar reasons introduced for lung microbiomes. Nevertheless, the ease of access still renders airway/oral microbes ideal candidates for lung cancer biomarkers. The most commonly examined samples were obtained from the oral cavity or bronchoalveolar lavage fluid (BALF).
The oral cavity harbors the second largest microbial community in our body; therefore, oral microbiomes possess a rich repertoire of microbes for biomarker identification. Oral samples can be obtained via oral wash [28,51,73] or saliva [38,57,60,61,67,68,71,78,85]. Other than oral samples, another easily accessible sampling site/biospecimen is sputum [25,27,37,42,46,49,50,58,60,63,69,76,87] or posterior oropharynx [21,62]; the sputum is the second most-used sample for lung cancer-associated microbiome studies. In an analysis of potential biomarkers for NSCLC metastasis using sputum and gut microbiomes, Lu et al. [42] suggested that several microbial biomarkers are shared between the sputum and gut, and the prediction power of sputum microbial biomarkers is similar to that of the combination of the sputum and gut. This further supports the use of sputum microbiome in lung cancer prognosis.
Other than the mouth and upper respiratory tract, the lower respiratory tract (other than the lung) is another popular (and mainly studied) sampling site for lung cancer-associated microbiome studies. Such samples can be acquired chiefly via BALF, although bronchial washing fluid and broncho epithelial brushing were adopted as well (refer to Table 1 as there are too many studies). The BALF sample has several unique advantages, including the relative ease of access, proximity to lung tissues, lower invasiveness and less ethical problems (especially when compared to healthy lung tissue samples). Moreover, although the lower respiratory tract may possess a unique microbiome (compared to the microbiome in other sites) [57,60,61], several independent studies reported that BALF or bronchial washing fluid is one of the most representative biospecimens for lung cancer-associated microbiome study among different sampling sites of the respiratory tract. Huang et al. [63] analyzed bronchial washing fluids and sputum samples from lung cancer patients and found that the microbiome of bronchial washing fluids was more similar to that of lung cancer tissues. They further observed that microbial biomarkers observed in bronchial washing fluid were more significantly associated with the different stages and subtypes of lung cancers (compared to those of sputum samples), indicating that bronchial fluid samples reflect lung cancer-specific microbes better. In addition, another study reported that the lung cancer-specific microbes detected in BALF mostly cover those identified in lung cancer tissues [32], further indicating that bronchial samples are an ideal alternative to lung tissues for lung cancer-associated microbiome study.
4. Frequently Altered Bacterial Genera in Lung Cancer Patients
While different lung microbiome profilings provided inconsistent results, certain bacteria are still frequently identified as lung cancer-specific (regardless of sampling sites), and their roles in lung cancer tumorigenesis have been elucidated. Therefore, these bacteria are promising lung cancer biomarkers and will be discussed in the following sections.
4.1. Veillonella
Although being a genus highly dominated in the airways and oral cavity [21,33], Veillonella in the respiratory tract or mouth is still perhaps one of the most identified microbial lung cancer biomarkers. Currently, there are 10 studies indicating that the abundance of Veillonella at various sites of the respiratory tract (e.g., sputum, saliva and BALF) is highly associated with lung cancers (or poor prognosis and distant metastasis of lung cancers, regardless of tumor subtypes), even the controls used were different [21,34,38,49,52,63,67,74,77,78]. For example, by analyzing airway brushing samples from lung cancer patients, non-cancer patients and healthy controls, Tsay et al. [74] found that oral taxa Veillonella is only highly enriched in lung cancer patients. The same group further analyzed the microbiome of lower airway brushing, transcriptomic data and the clinical data of 83 lung cancer patients (some transcriptomic data and clinical data were missing) and observed that Veillonella is highly enriched in patients with poor prognosis and tumor progression [52]. Similarly, an analysis of BALF samples [77] or salivary samples [78] demonstrated that Veillonella could be a potential biomarker of lung cancers. Interestingly, although Veillonella is the dominant genus in the induced sputum of either NSCLC patients or patients with chronic obstructive pulmonary disease (COPD), its abundance is decreased in patients with both NSCLC and COPD [27]. The authors proposed that the latter cases might be the conditions of cancer progression driven by COPD-induced inflammation, and the decreased Veillonella results from stress tolerance.
Due to the prevalence of Veillonella in lung cancer, the potential mechanism of Veillonella in lung cancer progression has also been studied. It has been shown that the upregulation of extracellular signal-regulated kinase (ERK) and phosphoinositide 3-kinase (PI3K) signaling pathways in lower airway transcriptomes of lung cancer patients is significantly associated with the enrichment of Veillonella in the same location [74]. Moreover, the in vitro co-culture of the Veillonella supernatant with airway epithelial cells leads to the activation of the PI3K pathway and the upregulation of inflammasome-related genes (e.g., IL-17) [74]. It has been shown that the PI3K pathway activation in the airway epithelium contributes to lung cancer initiation and progression [88,89], suggesting Veillonella’s potential carcinogenic role in lung cancer. In the subsequent study from the same group, the authors further confirmed this association using a KP mice model of lung cancer (with conditional activatable oncogenic Kras and Trp53 mutation, the mouse homolog of TP53). The intra-tracheal inoculation of Veillonella parvula to KP mice leads to lung dysbiosis and induces the strongest lower airway inflammation among all potential microbial biomarkers [52]. This results from the increased recruitment of Th17 and neutrophils, enhanced IL-17 production and an elevated expression of PD-1 in T cells [52]. Another study also reported that the systemic inflammation marker, neutrophil-lymphocyte ratio, is positively associated with the abundance of Veillonella in the saliva of NSCLC patients [67]. Zeng et al. [34] reported that Veillonella significantly promotes lung cancer progression in C57BL/6 mice as well. It should be noted that the association between PD-1+ T cells and Veillonella may make Veillonella a potential predictor of anti-PD-1 immunotherapy responses, as Veillonella dispar is dominated in the BALF of lung cancer patients with high PD-L1 expression [40]. These patients are also responders to immunotherapy. In conclusion, Veillonella can promote lung cancer progression by creating a pro-cancer immune microenvironment.
Notably, besides directly regulating host cells, Veillonella can also modulate the microbial community. In the murine model with CT26 colon carcinoma cells, Veillonella can significantly increase the abundance of pro-inflammatory bacteria Pseudomonas aeruginosa in tumor tissues. Increased Pseudomonas aeruginosa leads to increased blood TNF-α and poor survival outcomes [90]. In lung cancer, Pseudomonas is enriched in BALF [22], lower airway tract [52], cancer tissues [47,54] or the sputum of patients with distant metastasis as well [42]. The increased abundance of Pseudomonas in lung cancers may result from enriched Veillonella. Therefore, Veillonella may potentially modulate other lung cancer-associated microbes (e.g., Pseudomonas) and indirectly contribute to lung cancer progression.
4.2. Prevotella
Similarly to Veillonella, Prevotella is a genus that is highly enriched in airways [21,33]. There are currently nine studies showing Prevotella as a potential microbial biomarker for lung cancers [21,29,32,34,38,45,52,72,74]. Interestingly, Prevotella seems to be highly associated with Veillonella, as five studies observed that both Prevotella and Veillonella are enriched in lung cancer patients (although the correlation between Prevotella and lung cancer is not as significant as that for Veillonella) [21,34,38,52,74]. A typical healthy and balanced oral or lung microbiome consists of genera, including Streptococcus, Neisseria Prevotella, Veillonella, Porphyromonas and Fusobacterium [91]. It is possible that pathogenic events during lung cancer progression affect both Prevotella and Veillonella at the same time. However, the possibility of opportunistic infections cannot be excluded. Similarly to Veillonella, the co-culture of Prevotella with airway epithelial cells also leads to the upregulation of ERK and PI3K signaling pathways [74]. Therefore, Veillonella and Prevotella likely play similar roles in lung cancer progression.
4.3. Streptococcus
Perhaps Streptococcus is the most commonly detected genus across all lung cancer-associated microbiome profilings (using various samples). Over 20 studies demonstrate that changes in Streptococcus abundance are related to lung cancer (Table 1). While inconsistency exists, most of these studies reported that the abundance of Streptococcus positively correlates with lung cancer progression. Mechanistically, Streptococcus leads to the upregulation of ERK and PI3K signaling pathways in airway epithelial cells, which is the same as Prevotella and Veillonella [74]. In addition, it has been proposed that Streptococcus mitis can induce inflammation, Th17 activation and PD-L1 expression, leading to tumorigenesis (similarly to Veillonella) [57]. However, such chronic lung inflammation is potentially caused by lung dysbiosis; whether Streptococcus plays a vital role in it remains to be determined. Moreover, as Streptococcus is one of the most abundant genera in the respiratory tract [91], the enrichment of Streptococcus in lung cancer patients may result from opportunistic infection. Its presence in lung tissue may be a result of microaspirations from the oral cavity [57]. There are similar concerns for the other aforementioned dominant genera in the respiratory tract (i.e., Prevotella and Veillonella). However, the contributing roles of Veillonella in tumor progression have been elucidated, suggesting that it is not an opportunistic pathogen. While the precise role of Streptococcus in lung cancer remains to be determined, the increased abundance of Streptococcus can still be potentially used as a biomarker for immune dysregulation and lung cancer.
4.4. Acidovorax
Other than Veillonella, another commonly identified microbial lung cancer biomarker with potential mechanisms studied is Acidovorax. As introduced earlier, Acidovorax is a PAH-degrading microbe specifically associated with the LUSC of smokers [39,73]. Besides these two studies, other studies also reported that Acidovorax is enriched in LUSC tissues [49], and another targeted analysis of Acidovorax revealed that the abundance of Acidovorax is significantly elevated in LUSC patients with COPD or patients with LUSC relapse after surgery compared to patients with other conditions [92]. Interestingly, these studies also reported that the presence of Acidovorax is highly associated with TP53 mutations in lung cancers [39,73,92].
As a PAH-degrading bacterium, the enrichment of Acidovorax in the respiratory tract may result from environmental pressure from smoking or pollution. It is widely known that smoking directly leads to immune dysfunction and damages epithelial cells [93]. Acidovorax can protect these damaged cells by degrading toxic smoke compounds. Therefore, these damaged cells survive and contribute to potential tumorigenesis. Moreover, these pollutant-detoxication microbes have metabolic advantages and may form biofilms. Biofilm can potentially damage epithelial layers, leading to further bacterial invasion, DNA damage and chronic inflammation [94].
Furthermore, preliminary data suggested that Acidovorax temperans suppress both innate and adaptive immunity [95]. The phagocytosis of M2 macrophages is decreased after Acidovorax temperans engulfment, and these M2 macrophages can suppress T cell function via CD47-SIRPα immune checkpoints. Therefore, the nasal delivery of Acidovorax temperans accelerates lung tumorigenesis in the KPC murine models of LUAD driven by lung-specific Ad-Cre-activated Kras and Trp53 mutations [95]. The same group also observed that Acidovorax temperans’ exposure in murine lung cancer models increases pro-inflammatory cells (mostly neutrophils), CD4+ T cells and CD4– CD8– T cells (including RORγt+ IL17+ T cells). These results suggested that Acidovorax temperans can reshape the immune microenvironment of lung cancer and promote tumorigenesis [96].
4.5. Haemophilus
There are 11 independent lung cancer-associated microbiome sequencing studies [21,25,32,36,38,42,50,52,58,64,81] and one traditional culture-dependent study [97] demonstrating the association between Haemophilus enrichment (at various sites) and lung cancer (although such an association was not as significant as those between lung cancer and Veillonella). The commonly identified species of Haemophilus include Haemophilus parainfluenzae and Haemophilus influenzae, but Haemophilus haemolyticus is reported as well. However, the predictive value of Haemophilus abundance in lung cancer may require further validation, as there are studies indicating contradictory results: Haemophilus is only enriched in paracancerous normal tissue [49] or in the BALF of patients with COPD [45]. Such inconsistent observations are most likely a result of the fact that the Haemophilus genus is a ubiquitous bacterium that can be found in the respiratory tract of nearly 80% of healthy individuals [98]. Therefore, Haemophilus may be an opportunistic pathogen in lung cancer patients, as their immune systems are compromised.
4.6. Capnocytophaga
Most Capnocytophaga bacteria are normal bacteria commonly found in the oral cavity or oropharyngeal tracts of humans. However, under immune-compromised conditions, such as lung cancer, Capnocytophaga can be an opportunistic pathogen. Indeed, six studies reported an elevated Capnocytophaga abundance in the salivary, sputum, BALF and tumor tissues of lung cancer patients [19,44,49,55,65,78]. However, although the enrichment of Capnocytophaga in lung cancer may result from opportunistic infection, studies also reported that Capnocytophaga contributes to lung abscess formation [99]. Therefore, it is still possible that Capnocytophaga induces long-term inflammation, ultimately leading to tumor formation.
4.7. Other Commonly Identified Lung Cancer-Associated Microbes
The association between Acinetobacter and lung cancers has been reported extensively. However, there is enormous inconsistency across the studies: Five studies indicated that Acinetobacter was elevated in lung cancer patients [29,43,65,76,81], while four other studies reported that the abundance of Acinetobacter decreased in the same condition [27,36,71,72]. Such a lack of consensus occurs at the species level as well. This suggests that the variation in Acinetobacter abundances in lung cancer patients may result from microbiome dysbiosis, and Acinetobacter does not play an essential role in tumorigenesis. Similarly, contradictory results have been observed for Staphylococcus (Table 1). A further analysis should be performed at the species level (where many studies failed to do so). In addition, several studies have also reported an increased abundance of Rothia in lung cancer patients (Table 1). However, few papers suggested it can be a promising biomarker.
5. Conclusions
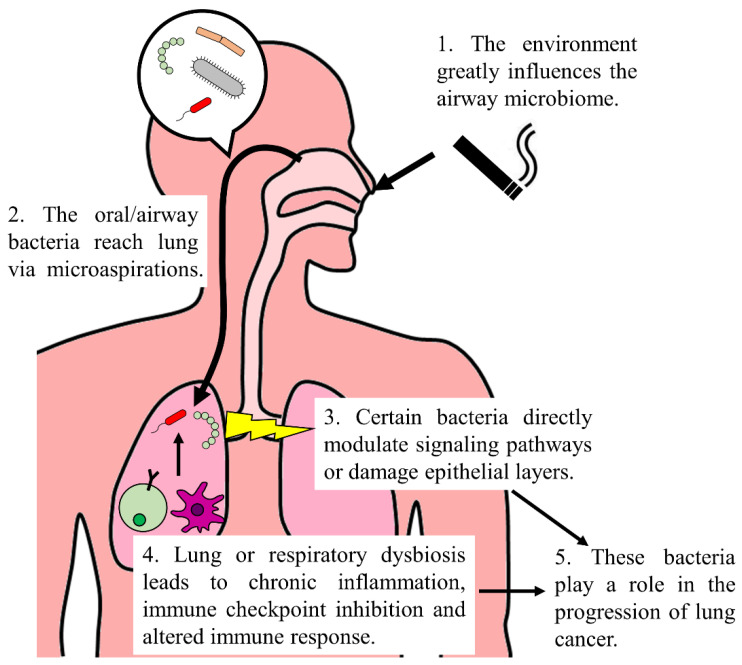
According to the WHO, lung cancer is the second most common cancer and the global leading cause of cancer-related death [14]. While the precise mechanisms require further elucidation, the microbiome undoubtedly plays an essential role in lung cancer initiation and progression (Figure 1). Therefore, numerous researchers have identified potential microbial biomarkers for lung cancers. However, a lack of consensus still exists across these studies. Some critical issues in this field still need to be addressed: Firstly, there are technical problems and variations in the studies’ designs. The collection of lung cancer-related microbiomes is challenging, and they are sensitive to contamination. The detection methods and controls used in each study vary as well. Initiating an international, multi-center, extensive cohort analysis (with standardized methodology) to identify potential microbial biomarkers of lung cancer thoroughly would be ideal. However, only one international, multi-center, extensive cohort analysis is not enough, because the second issue that needs addressing is the environmental influence on the respiratory tract’s microbiome. Therefore, regional analyses are still required to establish a local panel of biomarkers. Thirdly, lung/respiratory microbiome dysbiosis (instead of a specific bacterial species) seems to be a driving force in lung cancer formation. With the development of multi-omics and spatial transcriptomics analysis, the exact interaction between different microbes and cell types can be revealed. Lastly, the roles of other microbes (i.e., fungi and viruses, other than bacteria) in lung cancer remain unknown. Recently, a pan-cancer mycobiome analysis (using TCGA data) revealed that Blastomyces are enriched in lung cancer tissues [100]. A preliminary assessment of virome in the cancer tissue of LUAD also showed that lung cancer might possess unique virus compositions [101]. Hence, fungi and viruses can also serve as promising biomarkers for lung cancer, and further profilings are required.
Figure 1.
Involvement of the microbiome in lung cancer. The environmental factors (e.g., smoking) are the primary cause of variations in oral and airway microbiomes. These bacteria reach lung via microaspirations. Certain bacteria, like Veillonella and Acidovorax, can directly upregulate ERK and PI3K signaling pathways or damage epithelial cells. Moreover, lung or respiratory dysbiosis reshapes immune response, leading to aggravated Th17 and neutrophil responses, chronic inflammation and elevated levels of immune checkpoint molecules. Taken together, these microorganisms contribute to lung cancer development.
While there are still some critical problems and substantial inconsistencies in all lung cancer-associated microbiomes across the studies, a consensus is still reached with respect to the association between certain bacteria genera (such as Veillonella, Acidovorax, Prevotella, Streptococcus, Haemophilus and Capnocytophage) and lung cancer. While not all lung cancer-associated microbiome profilings detect the enrichment of these genera in lung cancers, the abundance of these genera is only increased in lung cancer patients. Therefore, these genera can serve as risk factors for lung cancer. Due to environmental variations and patient dissimilarities, obtaining globally accepted microbial biomarkers for lung cancer may be impossible. However, the microbiome studies of the respiratory tract still identified numerous microbes as risk factors. This can significantly help the diagnosis and prognosis of lung cancer patients.
Acknowledgments
We acknowledge our colleagues in our department for their ongoing advice. We apologize to the colleagues whose work we could not cite due to space limitations.
Author Contributions
Conceptualization, J.H. (Jiawen Huang), and J.H. (Juan Huang); investigation, J.H. (Jiawen Huang); writing—original draft, J.H. (Jiawen Huang); writing—review and editing, J.H. (Jiawen Huang) and J.H. (Juan Huang); supervision, J.H. (Juan Huang). All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was financially supported by the National Natural Science Foundation of China (NSFC 81500173).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sender R., Fuchs S., Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Grice E.A., Segre J.A. The human microbiome: Our second genome. Annu. Rev. Genom. Hum. Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Y., Pedersen O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021;19:55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 4.Hou K., Wu Z.X., Chen X.Y., Wang J.Q., Zhang D., Xiao C., Zhu D., Koya J.B., Wei L., Li J., et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022;7:135. doi: 10.1038/s41392-022-00974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yagi K., Huffnagle G.B., Lukacs N.W., Asai N. The Lung Microbiome during Health and Disease. Int. J. Mol. Sci. 2021;22:10872. doi: 10.3390/ijms221910872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch S.V., Pedersen O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert J.A., Blaser M.J., Caporaso J.G., Jansson J.K., Lynch S.V., Knight R. Current understanding of the human microbiome. Nat. Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullin N., Azevedo Antunes C., Straussman R., Stein-Thoeringer C.K., Elinav E. Microbiome and cancer. Cancer Cell. 2021;39:1317–1341. doi: 10.1016/j.ccell.2021.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Wroblewski L.E., Peek R.M., Jr., Wilson K.T. Helicobacter pylori and gastric cancer: Factors that modulate disease risk. Clin. Microbiol. Rev. 2010;23:713–739. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullman S., Pedamallu C.S., Sicinska E., Clancy T.E., Zhang X., Cai D., Neuberg D., Huang K., Guevara F., Nelson T., et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nejman D., Livyatan I., Fuks G., Gavert N., Zwang Y., Geller L.T., Rotter-Maskowitz A., Weiser R., Mallel G., Gigi E., et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu A., Yao B., Dong T., Chen Y., Yao J., Liu Y., Li H., Bai H., Liu X., Zhang Y., et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. 2022;185:1356–1372.e1326. doi: 10.1016/j.cell.2022.02.027. [DOI] [PubMed] [Google Scholar]
- 13.Kalaora S., Nagler A., Nejman D., Alon M., Barbolin C., Barnea E., Ketelaars S.L.C., Cheng K., Vervier K., Shental N., et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature. 2021;592:138–143. doi: 10.1038/s41586-021-03368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO Cancer. [(accessed on 23 October 2022)]. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer.
- 15.Dickson R.P., Erb-Downward J.R., Martinez F.J., Huffnagle G.B. The Microbiome and the Respiratory Tract. Annu. Rev. Physiol. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Dwyer D.N., Dickson R.P., Moore B.B. The Lung Microbiome, Immunity, and the Pathogenesis of Chronic Lung Disease. J. Immunol. 2016;196:4839–4847. doi: 10.4049/jimmunol.1600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huffnagle G.B., Dickson R.P., Lukacs N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal Immunol. 2017;10:299–306. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren L., Zhang R., Rao J., Xiao Y., Zhang Z., Yang B., Cao D., Zhong H., Ning P., Shang Y., et al. Transcriptionally Active Lung Microbiome and Its Association with Bacterial Biomass and Host Inflammatory Status. mSystems. 2018;3:e00199-18. doi: 10.1128/mSystems.00199-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y., Huang Y., Ding X., Yang Z., He L., Ning M., Yang Z., He D., Yang L., Liu Z., et al. A Multi-Omics Study of Familial Lung Cancer: Microbiome and Host Gene Expression Patterns. Front. Immunol. 2022;13:827953. doi: 10.3389/fimmu.2022.827953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong-Rolle A., Dong Q., Zhu Y., Divakar P., Hor J.L., Kedei N., Wong M., Tillo D., Conner E.A., Rajan A., et al. Spatial meta-transcriptomics reveal associations of intratumor bacteria burden with lung cancer cells showing a distinct oncogenic signature. J. Immunother. Cancer. 2022;10:e004698. doi: 10.1136/jitc-2022-004698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinhold L., Mollering A., Wallis S., Palade E., Schafer K., Dromann D., Rupp J., Graspeuntner S., Dalhoff K. Dissimilarity of Airway and Lung Tissue Microbiota in Smokers undergoing Surgery for Lung Cancer. Microorganisms. 2020;8:794. doi: 10.3390/microorganisms8060794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng L., Sun R., Zhu Y., Li Z., She X., Jian X., Yu F., Deng X., Sai B., Wang L., et al. Lung microbiome alterations in NSCLC patients. Sci. Rep. 2021;11:11736. doi: 10.1038/s41598-021-91195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G., Gail M.H., Consonni D., Carugno M., Humphrys M., Pesatori A.C., Caporaso N.E., Goedert J.J., Ravel J., Landi M.T. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17:163. doi: 10.1186/s13059-016-1021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan X., Wang Z., Li C., Lv K., Tian G., Tang M., Ji L., Yang J. Bacterial biomarkers capable of identifying recurrence or metastasis carry disease severity information for lung cancer. Front. Microbiol. 2022;13:1007831. doi: 10.3389/fmicb.2022.1007831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baranova E., Druzhinin V., Matskova L., Demenkov P., Volobaev V., Minina V., Larionov A., Titov V. Sputum Microbiome Composition in Patients with Squamous Cell Lung Carcinoma. Life. 2022;12:1365. doi: 10.3390/life12091365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Z., Tang J., Zhuang R., Meng D., Zhang L., Gu C., Teng X., Zhu Z., Liu J., Pang J., et al. The microbiome of lower respiratory tract and tumor tissue in lung cancer manifested as radiological ground-glass opacity. Front. Bioeng. Biotechnol. 2022;10:892613. doi: 10.3389/fbioe.2022.892613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J.Q., Chen Q., Wu S.J., Wang D.Q., Zhang S.Y., Zhang S.Z., Chen R.L., Wang J.F., Wang Z., Yu C.H. Potential Implications of the Lung Microbiota in Patients with Chronic Obstruction Pulmonary Disease and Non-Small Cell Lung Cancer. Front. Cell. Infect. Microbiol. 2022;12:937864. doi: 10.3389/fcimb.2022.937864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogtmann E., Hua X., Yu G., Purandare V., Hullings A.G., Shao D., Wan Y., Li S., Dagnall C.L., Jones K., et al. The oral microbiome and lung cancer risk: An analysis of 3 prospective cohort studies. J. Natl. Cancer Inst. 2022;114:1501–1510. doi: 10.1093/jnci/djac149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim O.H., Choi B.Y., Kim D.K., Kim N.H., Rho J.K., Sul W.J., Lee S.W. The microbiome of lung cancer tissue and its association with pathological and clinical parameters. Am. J. Cancer Res. 2022;12:2350–2362. [PMC free article] [PubMed] [Google Scholar]
- 30.Qian K., Deng Y., Krimsky W.S., Feng Y.G., Peng J., Tai Y.H., Peng H., Jiang L.H. Airway Microbiota in Patients With Synchronous Multiple Primary Lung Cancer: The Bacterial Topography of the Respiratory Tract. Front. Oncol. 2022;12:811279. doi: 10.3389/fonc.2022.811279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuhiro K., Tamiya M., Fujimoto K., Koyama S., Naito Y., Osa A., Hirai T., Suzuki H., Okamoto N., Shiroyama T., et al. Bronchoalveolar lavage fluid reveals factors contributing to the efficacy of PD-1 blockade in lung cancer. JCI Insight. 2022;7:e157915. doi: 10.1172/jci.insight.157915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M., Zhang Y., Han Y., Zhao X., Sun Y. Lung microbiota features of stage III and IV non-small cell lung cancer patients without lung infection. Transl. Cancer Res. 2022;11:426–434. doi: 10.21037/tcr-22-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall E.A., Filho F.S.L., Sin D.D., Lam S., Leung J.M., Lam W.L. Distinct bronchial microbiome precedes clinical diagnosis of lung cancer. Mol. Cancer. 2022;21:68. doi: 10.1186/s12943-022-01544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng W., Zhao C., Yu M., Chen H., Pan Y., Wang Y., Bao H., Ma H., Ma S. Alterations of lung microbiota in patients with non-small cell lung cancer. Bioengineered. 2022;13:6665–6677. doi: 10.1080/21655979.2022.2045843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu S., Cheng Z., Yin Z., Xu J., Wu F., Jin Y., Yang G. Airway Fusobacterium is Associated with Poor Response to Immunotherapy in Lung Cancer. Onco Targets Ther. 2022;15:201–213. doi: 10.2147/OTT.S348382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M., Zhang Y., Sun Y., Wang S., Liang H., Han Y. Intratumoral Microbiota Impacts the First-Line Treatment Efficacy and Survival in Non-Small Cell Lung Cancer Patients Free of Lung Infection. J. Healthc. Eng. 2022;2022:5466853. doi: 10.1155/2022/5466853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang D.H., He J., Su X.F., Wen Y.N., Zhang S.J., Liu L.Y., Zhao H., Ye C.P., Wu J.H., Cai S., et al. The airway microbiota of non-small cell lung cancer patients and its relationship to tumor stage and EGFR gene mutation. Thorac. Cancer. 2022;13:858–869. doi: 10.1111/1759-7714.14340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy P., Sarma A., Kataki A.C., Rai A.K., Chattopadhyay I. Salivary microbial dysbiosis may predict lung adenocarcinoma: A pilot study. Indian J. Pathol. Microbiol. 2022;65:123–128. doi: 10.4103/IJPM.IJPM_1111_20. [DOI] [PubMed] [Google Scholar]
- 39.Dong H., Tan Q., Xu Y., Zhu Y., Yao Y., Wang Y., Li C., Li H., Zhang G., Xiong Y., et al. Convergent alteration of lung tissue microbiota and tumor cells in lung cancer. iScience. 2022;25:103638. doi: 10.1016/j.isci.2021.103638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jang H.J., Choi J.Y., Kim K., Yong S.H., Kim Y.W., Kim S.Y., Kim E.Y., Jung J.Y., Kang Y.A., Park M.S., et al. Relationship of the lung microbiome with PD-L1 expression and immunotherapy response in lung cancer. Respir. Res. 2021;22:322. doi: 10.1186/s12931-021-01919-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boesch M., Baty F., Albrich W.C., Flatz L., Rodriguez R., Rothschild S.I., Joerger M., Fruh M., Brutsche M.H. Local tumor microbial signatures and response to checkpoint blockade in non-small cell lung cancer. Oncoimmunology. 2021;10:1988403. doi: 10.1080/2162402X.2021.1988403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu H., Gao N.L., Tong F., Wang J., Li H., Zhang R., Ma H., Yang N., Zhang Y., Wang Y., et al. Alterations of the Human Lung and Gut Microbiomes in Non-Small Cell Lung Carcinomas and Distant Metastasis. Microbiol. Spectr. 2021;9:e0080221. doi: 10.1128/Spectrum.00802-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang Y.S., Hsu M.H., Tu S.J., Yen J.C., Lee Y.T., Fang H.Y., Chang J.G. Metatranscriptomic Analysis of Human Lung Metagenomes from Patients with Lung Cancer. Genes. 2021;12:1458. doi: 10.3390/genes12091458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi J., Yang Y., Xie H., Wang X., Wu J., Long J., Courtney R., Shu X.O., Zheng W., Blot W.J., et al. Association of oral microbiota with lung cancer risk in a low-income population in the Southeastern USA. Cancer Causes Control. 2021;32:1423–1432. doi: 10.1007/s10552-021-01490-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seixas S., Kolbe A.R., Gomes S., Sucena M., Sousa C., Vaz Rodrigues L., Teixeira G., Pinto P., Tavares de Abreu T., Barbara C., et al. Comparative analysis of the bronchoalveolar microbiome in Portuguese patients with different chronic lung disorders. Sci. Rep. 2021;11:15042. doi: 10.1038/s41598-021-94468-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C., Wang J., Sun Z., Cao Y., Mu Z., Ji X. Commensal microbiota contributes to predicting the response to immune checkpoint inhibitors in non-small-cell lung cancer patients. Cancer Sci. 2021;112:3005–3017. doi: 10.1111/cas.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dumont-Leblond N., Veillette M., Racine C., Joubert P., Duchaine C. Non-small cell lung cancer microbiota characterization: Prevalence of enteric and potentially pathogenic bacteria in cancer tissues. PLoS ONE. 2021;16:e0249832. doi: 10.1371/journal.pone.0249832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma Y., Qiu M., Wang S., Meng S., Yang F., Jiang G. Distinct tumor bacterial microbiome in lung adenocarcinomas manifested as radiological subsolid nodules. Transl. Oncol. 2021;14:101050. doi: 10.1016/j.tranon.2021.101050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leng Q., Holden V.K., Deepak J., Todd N.W., Jiang F. Microbiota Biomarkers for Lung Cancer. Diagnostics. 2021;11:407. doi: 10.3390/diagnostics11030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Druzhinin V.G., Matskova L.V., Demenkov P.S., Baranova E.D., Volobaev V.P., Minina V.I., Larionov A.V., Titov V.A., Fucic A. Genetic damage in lymphocytes of lung cancer patients is correlated to the composition of the respiratory tract microbiome. Mutagenesis. 2021;36:143–153. doi: 10.1093/mutage/geab004. [DOI] [PubMed] [Google Scholar]
- 51.Hosgood H.D., Cai Q., Hua X., Long J., Shi J., Wan Y., Yang Y., Abnet C., Bassig B.A., Hu W., et al. Variation in oral microbiome is associated with future risk of lung cancer among never-smokers. Thorax. 2021;76:256–263. doi: 10.1136/thoraxjnl-2020-215542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsay J.J., Wu B.G., Sulaiman I., Gershner K., Schluger R., Li Y., Yie T.A., Meyn P., Olsen E., Perez L., et al. Lower Airway Dysbiosis Affects Lung Cancer Progression. Cancer Discov. 2021;11:293–307. doi: 10.1158/2159-8290.CD-20-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhuo M., An T., Zhang C., Wang Z. Characterization of Microbiota in Cancerous Lung and the Contralateral Non-Cancerous Lung Within Lung Cancer Patients. Front. Oncol. 2020;10:1584. doi: 10.3389/fonc.2020.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovaleva O., Podlesnaya P., Rashidova M., Samoilova D., Petrenko A., Zborovskaya I., Mochalnikova V., Kataev V., Khlopko Y., Plotnikov A., et al. Lung Microbiome Differentially Impacts Survival of Patients with Non-Small Cell Lung Cancer Depending on Tumor Stroma Phenotype. Biomedicines. 2020;8:349. doi: 10.3390/biomedicines8090349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng C., Wang Z., Wang J., Ding C., Sun C., Liu P., Xu X., Liu Y., Chen B., Gu B. Characterization of the lung microbiome and exploration of potential bacterial biomarkers for lung cancer. Transl. Lung Cancer Res. 2020;9:693–704. doi: 10.21037/tlcr-19-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao Q., Ma W., Wang Z., Liang Y., Zhang T., Yang Y., Xia W., Jiang F., Hu J., Xu L. Differential flora in the microenvironment of lung tumor and paired adjacent normal tissues. Carcinogenesis. 2020;41:1094–1103. doi: 10.1093/carcin/bgaa044. [DOI] [PubMed] [Google Scholar]
- 57.Bello S., Vengoechea J.J., Ponce-Alonso M., Figueredo A.L., Minchole E., Rezusta A., Gambo P., Pastor J.M., Javier G., Del Campo R. Core Microbiota in Central Lung Cancer With Streptococcal Enrichment as a Possible Diagnostic Marker. Arch. Bronconeumol. 2021;57:681–689. doi: 10.1016/j.arbres.2020.05.034. [DOI] [PubMed] [Google Scholar]
- 58.Druzhinin V.G., Matskova L.V., Demenkov P.S., Baranova E.D., Volobaev V.P., Minina V.I., Apalko S.V., Churina M.A., Romanyuk S.A., Shcherbak S.G., et al. Taxonomic diversity of sputum microbiome in lung cancer patients and its relationship with chromosomal aberrations in blood lymphocytes. Sci. Rep. 2020;10:9681. doi: 10.1038/s41598-020-66654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong L.M., Shende N., Li W.T., Castaneda G., Apostol L., Chang E.Y., Ongkeko W.M. Comparative Analysis of Age- and Gender-Associated Microbiome in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. Cancers. 2020;12:1447. doi: 10.3390/cancers12061447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bingula R., Filaire E., Molnar I., Delmas E., Berthon J.Y., Vasson M.P., Bernalier-Donadille A., Filaire M. Characterisation of microbiota in saliva, bronchoalveolar lavage fluid, non-malignant, peritumoural and tumour tissue in non-small cell lung cancer patients: A cross-sectional clinical trial. Respir. Res. 2020;21:129. doi: 10.1186/s12931-020-01392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patnaik S.K., Cortes E.G., Kannisto E.D., Punnanitinont A., Dhillon S.S., Liu S., Yendamuri S. Lower airway bacterial microbiome may influence recurrence after resection of early-stage non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2021;161:419–429.e416. doi: 10.1016/j.jtcvs.2020.01.104. [DOI] [PubMed] [Google Scholar]
- 62.Ekanayake A., Madegedara D., Chandrasekharan V., Magana-Arachchi D. Respiratory Bacterial Microbiota and Individual Bacterial Variability in Lung Cancer and Bronchiectasis Patients. Indian J. Microbiol. 2020;60:196–205. doi: 10.1007/s12088-019-00850-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang D., Su X., Yuan M., Zhang S., He J., Deng Q., Qiu W., Dong H., Cai S. The characterization of lung microbiome in lung cancer patients with different clinicopathology. Am. J. Cancer Res. 2019;9:2047–2063. [PMC free article] [PubMed] [Google Scholar]
- 64.Jin J., Gan Y., Liu H., Wang Z., Yuan J., Deng T., Zhou Y., Zhu Y., Zhu H., Yang S., et al. Diminishing microbiome richness and distinction in the lower respiratory tract of lung cancer patients: A multiple comparative study design with independent validation. Lung Cancer. 2019;136:129–135. doi: 10.1016/j.lungcan.2019.08.022. [DOI] [PubMed] [Google Scholar]
- 65.Gomes S., Cavadas B., Ferreira J.C., Marques P.I., Monteiro C., Sucena M., Sousa C., Vaz Rodrigues L., Teixeira G., Pinto P., et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci. Rep. 2019;9:12838. doi: 10.1038/s41598-019-49195-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren Y., Su H., She Y., Dai C., Xie D., Narrandes S., Huang S., Chen C., Xu W. Whole genome sequencing revealed microbiome in lung adenocarcinomas presented as ground-glass nodules. Transl. Lung Cancer Res. 2019;8:235–246. doi: 10.21037/tlcr.2019.06.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang W., Luo J., Dong X., Zhao S., Hao Y., Peng C., Shi H., Zhou Y., Shan L., Sun Q., et al. Salivary Microbial Dysbiosis is Associated with Systemic Inflammatory Markers and Predicted Oral Metabolites in Non-Small Cell Lung Cancer Patients. J. Cancer. 2019;10:1651–1662. doi: 10.7150/jca.28077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang K., Huang Y., Zhang Z., Liao J., Ding Y., Fang X., Liu L., Luo J., Kong J. A Preliminary Study of Microbiota Diversity in Saliva and Bronchoalveolar Lavage Fluid from Patients with Primary Bronchogenic Carcinoma. Med. Sci. Monit. 2019;25:2819–2834. doi: 10.12659/MSM.915332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hosgood H.D., 3rd, Mongodin E.F., Wan Y., Hua X., Rothman N., Hu W., Vermeulen R., Seow W.J., Rohan T., Xu J., et al. The respiratory tract microbiome and its relationship to lung cancer and environmental exposures found in rural china. Environ. Mol. Mutagen. 2019;60:617–623. doi: 10.1002/em.22291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peters B.A., Hayes R.B., Goparaju C., Reid C., Pass H.I., Ahn J. The Microbiome in Lung Cancer Tissue and Recurrence-Free Survival. Cancer Epidemiol. Biomarkers Prev. 2019;28:731–740. doi: 10.1158/1055-9965.EPI-18-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang J., Mu X., Wang Y., Zhu D., Zhang J., Liang C., Chen B., Wang J., Zhao C., Zuo Z., et al. Dysbiosis of the Salivary Microbiome Is Associated With Non-smoking Female Lung Cancer and Correlated With Immunocytochemistry Markers. Front. Oncol. 2018;8:520. doi: 10.3389/fonc.2018.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Y., O’Brien J.L., Ajami N.J., Scheurer M.E., Amirian E.S., Armstrong G., Tsavachidis S., Thrift A.P., Jiao L., Wong M.C., et al. Lung tissue microbial profile in lung cancer is distinct from emphysema. Am. J. Cancer Res. 2018;8:1775–1787. [PMC free article] [PubMed] [Google Scholar]
- 73.Greathouse K.L., White J.R., Vargas A.J., Bliskovsky V.V., Beck J.A., von Muhlinen N., Polley E.C., Bowman E.D., Khan M.A., Robles A.I., et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. doi: 10.1186/s13059-018-1501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsay J.J., Wu B.G., Badri M.H., Clemente J.C., Shen N., Meyn P., Li Y., Yie T.A., Lhakhang T., Olsen E., et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. Am. J. Respir. Crit. Care Med. 2018;198:1188–1198. doi: 10.1164/rccm.201710-2118OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu H.X., Tao L.L., Zhang J., Zhu Y.G., Zheng Y., Liu D., Zhou M., Ke H., Shi M.M., Qu J.M. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int. J. Cancer. 2018;142:769–778. doi: 10.1002/ijc.31098. [DOI] [PubMed] [Google Scholar]
- 76.Cameron S.J.S., Lewis K.E., Huws S.A., Hegarty M.J., Lewis P.D., Pachebat J.A., Mur L.A.J. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PLoS ONE. 2017;12:e0177062. doi: 10.1371/journal.pone.0177062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee S.H., Sung J.Y., Yong D., Chun J., Kim S.Y., Song J.H., Chung K.S., Kim E.Y., Jung J.Y., Kang Y.A., et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. 2016;102:89–95. doi: 10.1016/j.lungcan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 78.Yan X., Yang M., Liu J., Gao R., Hu J., Li J., Zhang L., Shi Y., Guo H., Cheng J., et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 2015;5:3111–3122. [PMC free article] [PubMed] [Google Scholar]
- 79.Hosgood H.D., 3rd, Sapkota A.R., Rothman N., Rohan T., Hu W., Xu J., Vermeulen R., He X., White J.R., Wu G., et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ. Mol. Mutagen. 2014;55:643–651. doi: 10.1002/em.21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chakravorty S., Helb D., Burday M., Connell N., Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods. 2007;69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang M., Zhang Y., Han Y., Zhao X., Sun Y. Characteristics of pathogenic microbes in lung microenvironment of lung cancer patients without respiratory infection. J. BUON. 2021;26:1862–1870. [PubMed] [Google Scholar]
- 82.Rodriguez R.M., Hernandez B.Y., Menor M., Deng Y., Khadka V.S. The landscape of bacterial presence in tumor and adjacent normal tissue across 9 major cancer types using TCGA exome sequencing. Comput. Struct. Biotechnol. J. 2020;18:631–641. doi: 10.1016/j.csbj.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Poore G.D., Kopylova E., Zhu Q., Carpenter C., Fraraccio S., Wandro S., Kosciolek T., Janssen S., Metcalf J., Song S.J., et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klein M., Pragman A.A., Wendt C. Biomarkers and the microbiome in the detection and treatment of early-stage non-small cell lung cancer. Semin. Oncol. 2022;49:285–297. doi: 10.1053/j.seminoncol.2022.06.011. [DOI] [PubMed] [Google Scholar]
- 85.Ying K.L., Brasky T.M., Freudenheim J.L., McElroy J.P., Nickerson Q.A., Song M.A., Weng D.Y., Wewers M.D., Whiteman N.B., Mathe E.A., et al. Saliva and Lung Microbiome Associations with Electronic Cigarette Use and Smoking. Cancer Prev. Res. 2022;15:435–446. doi: 10.1158/1940-6207.CAPR-21-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berger G., Wunderink R.G. Lung microbiota: Genuine or artifact? Isr. Med. Assoc. J. 2013;15:731–733. [PubMed] [Google Scholar]
- 87.Ran Z., Liu J., Wang F., Xin C., Shen X., Zeng S., Song Z., Xiong B. Analysis of Pulmonary Microbial Diversity in Patients with Advanced Lung Cancer Based on High-throughput Sequencing Technology. Zhongguo Fei Ai Za Zhi. 2020;23:1031–1038. doi: 10.3779/j.issn.1009-3419.2020.103.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scrima M., De Marco C., Fabiani F., Franco R., Pirozzi G., Rocco G., Ravo M., Weisz A., Zoppoli P., Ceccarelli M., et al. Signaling networks associated with AKT activation in non-small cell lung cancer (NSCLC): New insights on the role of phosphatydil-inositol-3 kinase. PLoS ONE. 2012;7:e30427. doi: 10.1371/journal.pone.0030427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gustafson A.M., Soldi R., Anderlind C., Scholand M.B., Qian J., Zhang X., Cooper K., Walker D., McWilliams A., Liu G., et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci. Transl. Med. 2010;2:26ra25. doi: 10.1126/scitranslmed.3000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pustelny C., Komor U., Pawar V., Lorenz A., Bielecka A., Moter A., Gocht B., Eckweiler D., Musken M., Grothe C., et al. Contribution of Veillonella parvula to Pseudomonas aeruginosa-mediated pathogenicity in a murine tumor model system. Infect. Immun. 2015;83:417–429. doi: 10.1128/IAI.02234-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kononen E., Gursoy U.K. Oral Prevotella Species and Their Connection to Events of Clinical Relevance in Gastrointestinal and Respiratory Tracts. Front. Microbiol. 2021;12:798763. doi: 10.3389/fmicb.2021.798763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shimizu M., Miyanaga A., Seike M., Matsuda K., Matsumoto M., Noro R., Fujita K., Mano Y., Furuya N., Kubota K., et al. The respiratory microbiome associated with chronic obstructive pulmonary disease comorbidity in non-small cell lung cancer. Thorac. Cancer. 2022;13:1940–1947. doi: 10.1111/1759-7714.14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Adar S.D., Huffnagle G.B., Curtis J.L. The respiratory microbiome: An underappreciated player in the human response to inhaled pollutants? Ann. Epidemiol. 2016;26:355–359. doi: 10.1016/j.annepidem.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dejea C.M., Fathi P., Craig J.M., Boleij A., Taddese R., Geis A.L., Wu X., DeStefano Shields C.E., Hechenbleikner E.M., Huso D.L., et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592–597. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Robles A.I., Cooks T., Vega-Valle E., Vetizou M., Rose U., Miyanaga A., Trehan A., Oike T., Ryan B.M., Sen S.J., et al. Abstract PR07: Role of the microbiota in inflammation and lung cancer. Clin. Cancer Res. 2018;24:PR07. doi: 10.1158/1557-3265.AACRIASLC18-PR07. [DOI] [Google Scholar]
- 96.Zhang C., von Muhlinen N., Romina A., Stone J., Goldszmid R., Harris C. Acidovorax temperans modulates the host immune microenvironment to promote lung cancer development. Cancer Res. 2021;81:1782. doi: 10.1158/1538-7445.AM2021-1782. [DOI] [Google Scholar]
- 97.Kosikowska U., Biernasiuk A., Rybojad P., Los R., Malm A. Haemophilus parainfluenzae as a marker of the upper respiratory tract microbiota changes under the influence of preoperative prophylaxis with or without postoperative treatment in patients with lung cancer. BMC Microbiol. 2016;16:62. doi: 10.1186/s12866-016-0679-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Middleton A.M., Dowling R.B., Mitchell J.L., Watanabe S., Rutman A., Pritchard K., Tillotson G., Hill S.L., Wilson R. Haemophilus parainfluenzae infection of respiratory mucosa. Respir. Med. 2003;97:375–381. doi: 10.1053/rmed.2002.1454. [DOI] [PubMed] [Google Scholar]
- 99.Thirumala R., Rappo U., Babady N.E., Kamboj M., Chawla M. Capnocytophaga lung abscess in a patient with metastatic neuroendocrine tumor. J. Clin. Microbiol. 2012;50:204–207. doi: 10.1128/JCM.05306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dohlman A.B., Klug J., Mesko M., Gao I.H., Lipkin S.M., Shen X., Iliev I.D. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell. 2022;185:3807–3822.e12. doi: 10.1016/j.cell.2022.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cai H.Z., Zhang H., Yang J., Zeng J., Wang H. Preliminary assessment of viral metagenome from cancer tissue and blood from patients with lung adenocarcinoma. J. Med. Virol. 2021;93:5126–5133. doi: 10.1002/jmv.26887. [DOI] [PubMed] [Google Scholar]