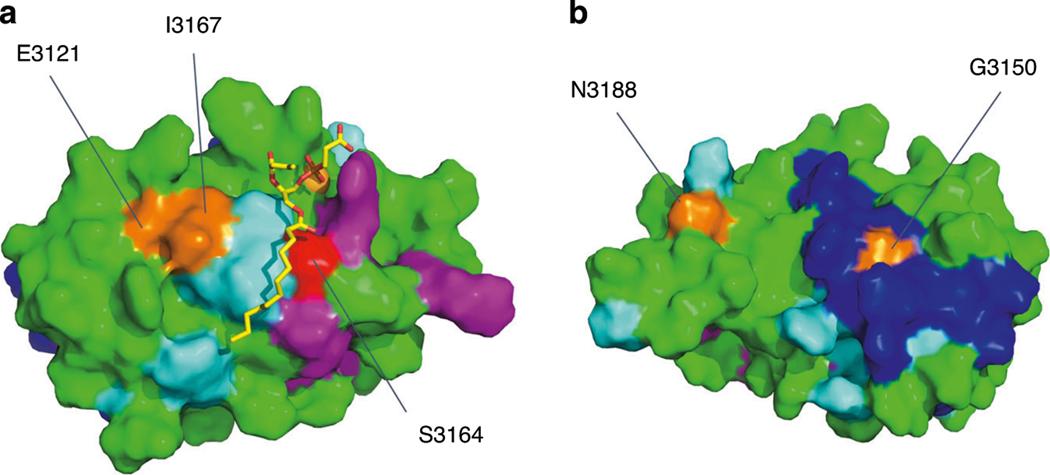
Fig. 3. Nuclear magnetic resonance (NMR) model of the PC1 PLAT domain with surface mapping of specific ligand-binding residues.
a Membrane-facing side showing phosphatidylserine, PS (stick), Ca2+ (orange sphere). Surface representation indicating residues binding PS (light blue) and β-arrestin1/2 (pink). The key Ser3164 residue (red) which modulates PI4P and β-arrestin1/2 binding is indicated.23 b Cytoplasmic-facing surface of PLAT showing a predicted protein interaction domain (deep blue). Several residues identified to be altered in this study are labeled (orange) and mapped onto this model, i.e., E3121, I3167, G3150, and N3188. Note the close proximity of E3121 and 13167 to the same Ca2+−dependent PS-binding pocket. E3121 also forms part of a functional YEIL3123 motif involved in AP2-mediated internalization of PC1.23.