Figure 2.
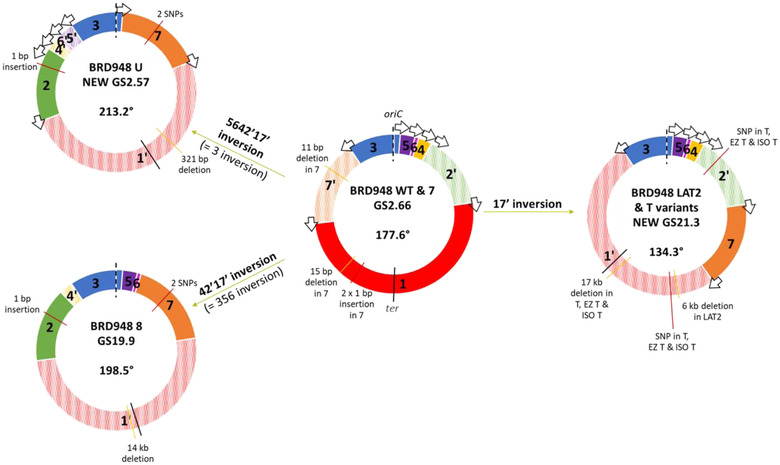
Genome rearrangements of variants relative to WT. Schematic showing variant genome structures (GSs) and the rearrangement of WT fragments required to achieve these. GS fragments are labeled in respect to the Salmonella enterica database reference LT2 (genome accession GCF_000006945.2) and drawn with oriC at the 12 o'clock position and working in a clockwise fashion around the chromosome. The fragment containing origin of replication (here fragment 3) has its orientation fixed to match the orientation of the database reference and therefore any inversion involving fragment 3 is depicted as the rest of the chromosome inverted. The depicted inversion is given in bold but for clarity the equivalent inversion is given in parentheses. Individual inverted fragment orientations are denoted prime (ʹ) with striped colors. Ori‐ter balance is given in degrees for each GS, going clockwise from ter to oriC as drawn. Arrows: ribosomal operons; oriC and dashed lines: origin of replication; and ter and black whole lines: terminus of replication. Data from genome sequencing are used to identify insertions (red lines) and deletions (yellow lines) in each variant in comparison to WT; bp, base pairs.
