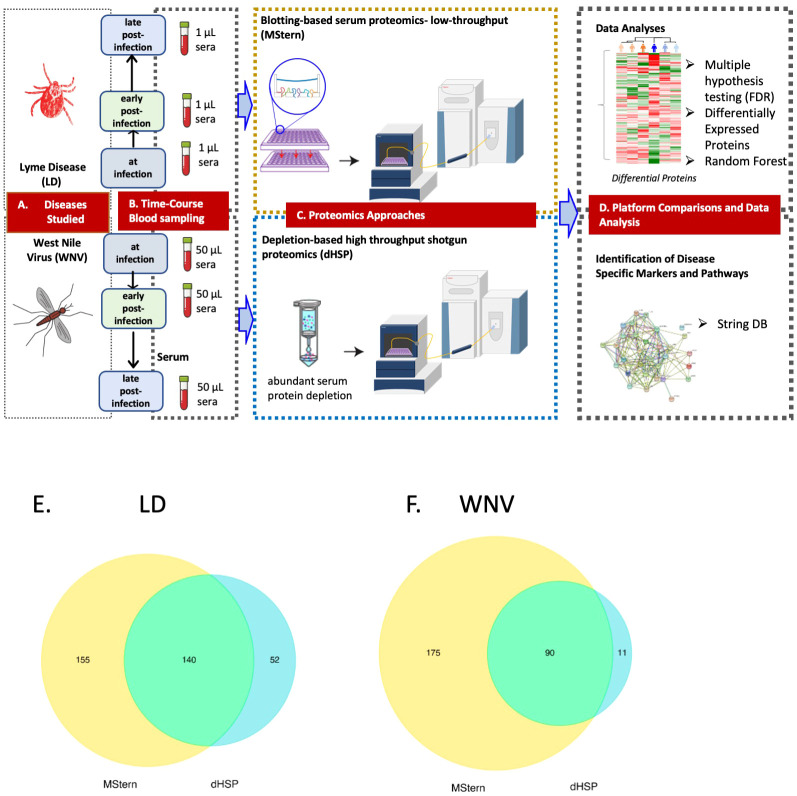
Figure 3.
LEGEND - Schematic representation of the overall study. (A) Diseases studied- Lyme disease (tick-borne) and West Nile Virus infection (mosquito-borne).(B) Blood collection by venipuncture and serum separation (C) Serum protein processing platforms– MStern spotting of one µl serum on PVDF microtiter membrane without depleting high abundant protein followed by protein denaturation, reduction and alkylation of cysteine residues, rapid protein digestion, LCMS (top panel) and dHSP with depletion of high abundance blood proteins using spin columns followed by protein denaturation, reduction and alkylation of cysteine residues, rapid protein digestion and LCMS shotgun proteomics (bottom panel) (D) Data analysis and data interrogation for marker discovery and underlying molecular mechanisms. Venn Diagram of the analyzed proteins from both the dHSP and MStern platforms in LD (E) and WNV (F). The optimized depletion-based method is an unbiased approach that was based on shotgun proteomics method that required 50 μl serum for depletion step and 1μg of tryptic peptides for LC/MS analysis. The MStern platform starts with 1μl serum on a 96 well-plate-based method with no depletion step required in WNV.