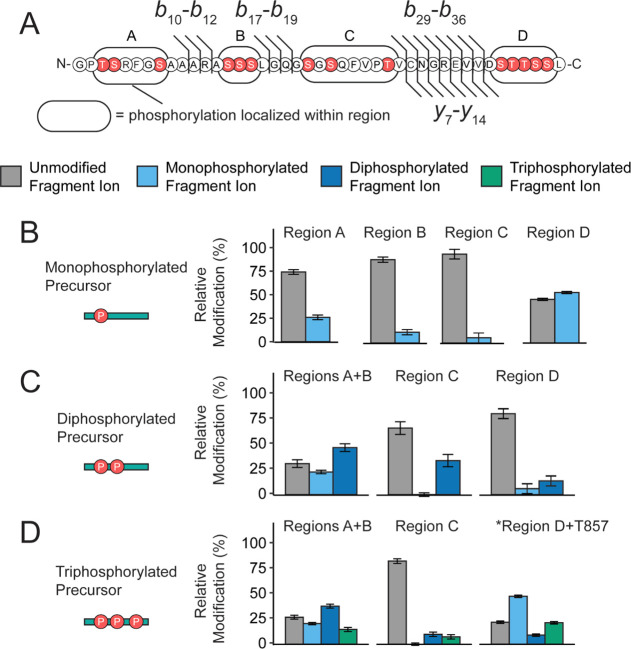
Figure 3.
mGluR2 is subject to basal isomeric phosphorylation states. (A) Sequence of mGluR2 cleavage product and potential phosphorylation sites (red) used for PRM studies, b-type and y-type fragment ions utilized for PRM studies are denoted as black flags. Fragment ion numbering is based on the number of residues in the cleavage product (P2 = PA830 in accession #Q14416-1). Phosphorylation content was localized between regions A, B, C, and D for the (B) mono-, (C) di-, (D) and triphosphorylated populations. The intact precursor corresponding to each set of plots is shown pictorially on the left. Color fill denotes the number of phosphorylations localized within a given region using fragment ions from the PRM assay. No phosphorylation is denoted with gray, monophosphorylation is denoted with light blue, diphosphorylation is denoted with dark blue, and triphosphorylation is denoted with green. For the higher stoichiometries, regions A and B were combined based on fragment ion coverage. *For the triphosphorylated population, y19 to y16 were used to calculate occupancy in region D + T857. Error bars represent the standard error of the mean (N = 3 biological replicates and 3 technical replicates).