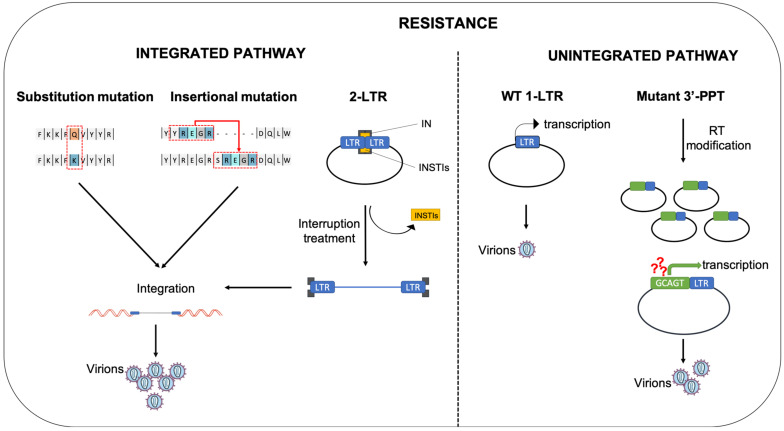
Figure 5.
Different pathways of INSTI resistance. Two main pathways involving the integration step or unintegrated viral DNA can be responsible for INSTI resistance. Substitution mutations (replacement of one amino-acid by another one) or insertional mutations (insertion of one or several amino-acids) in the integrase protein can lead to INSTI resistance. In these pathways, integration of the viral DNA can occur because the mutated integrase is resistant to INSTIs but still active to mediate integration. Integrated DNA then allows the production of new viruses bearing these resistance mutations. Blocking of integration by INSTI treatment leads to accumulation of unintegrated DNA, in particular, 2-LTR circles. In case of interruption of treatment, integration from 2-LTR circles can occur due to the specific cleavage of the LTR-LTR junction before integration after release of the INSTI compound from the IN/DNA complex. Thus, 2-LTRc accumulated in cells treated with an INSTI may constitute a reserve supply of HIV-1 genomes that could be integrated de novo and revive viral replication. Unintegrated viral DNA could also contribute to INSTI resistance by a basal production of viruses from 1-LTR circles. Furthermore, mutations in the 3′-PPT region modify the reverse transcription step. In this case, reverse transcription does not result in linear DNA but in a 1-LTR circle. One hypothesis is that the 3′-PPT mutation leads to a higher transcription efficiency leading a higher viral production compared to the production from a 1-LTR circle that does not highlight the 3′-PPT mutation. Viruses with 3′-PPT mutations are resistant to INSTIs since their replication is independent of the integration pathway. Yellow square: INSTI. Black arrow: basal transcription from 1-LTR circles coming from a WT infection. Green arrow: hypothesis of higher transcription due to the 3′-PPT mutation.