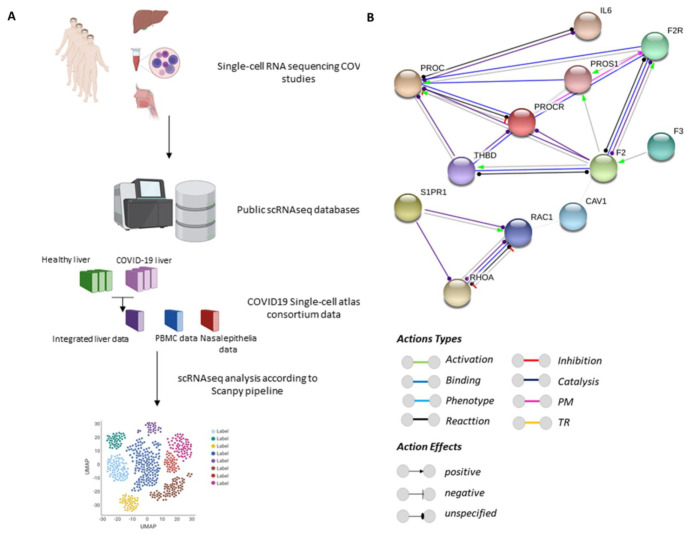
Figure 1.
Experimental design and protein–protein interaction network. (A). Public data from single-cell RNA sequencing (scRNA-seq) studies on the human tissues (liver, peripheral blood mononuclear cells (PBMC), and nasal epithelia) were retrieved from the website portal www.COVID-19cellatlas.org accessed on 16 April 2022 and Gene Expression Omnibus (GEO) accession numbers GSE171668 and GSE115469. (B). The PROC interactome was retrieved with the string-db data mining toolkit (https://version-11-0.string-db.org/cgi/network.pl?taskId=lu4XDH7F7cOZ, accessed on 16 April 2022). The sources used for functional interactions were neighborhood, experiments, gene fusion, databases, co-occurrence and co-expression, and visualized by the ‘molecular actions’. The network nodes represent proteins and the edges represent protein–protein associations. The edges indicate the known molecular action of a protein node relative to another protein node. PM: post-translational modification; TR: transcriptional regulation.