FIGURE 2.
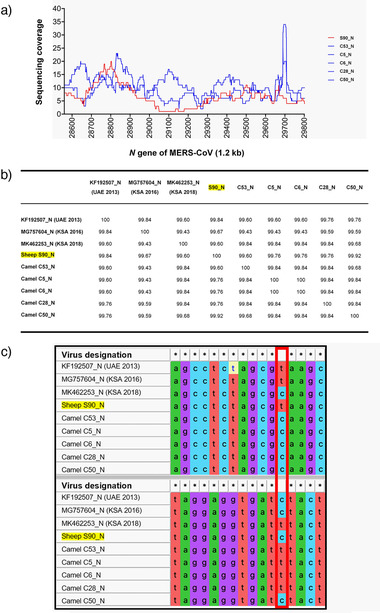
Next generation sequencing analysis of the MERS‐CoV N gene of sheep and camels. (a) Sequencing coverage of the MERS‐CoV N gene derived from one sheep (S90_N) and five camels (C53_N, C5_N, C6_N, C28_N, and C50_N). The average coverage of the N gene across all samples was >6X. X‐axis = N gene of MERS‐CoV (1.2 kb) and Y‐axis = sequencing coverage; red line = sheep‐derived MERS‐CoV, blue lines = dromedary‐derived MERS‐CoV. (b) Percentage of N gene sequence identity of the same samples along with three references MERS‐CoV sequences (GenBank acc. numbers KF192507, MG757604, and MK462253). The sample collected from the sheep showed high nucleotide identity (>99%) with all camel‐derived MERS‐CoV sequences as well as the reference sequences. (c) Multiple sequence alignment of the N gene of the same MERS‐CoV samples (1.2 kb). Two short representative regions with nucleotide sequence variations are shown: positions 533 and 637 of the N gene (highlighted by a red vertical box); conserved positions are indicated by asterisks (*). Multiple sequence alignment was performed using MAFFT version 7.455
