FIGURE 1.
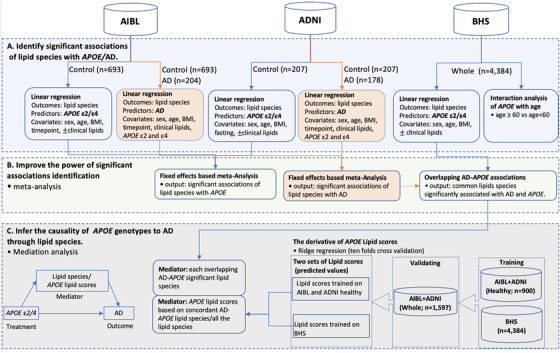
Study design. In this study, the analyses include three main sections: the identification of the significant associations of lipid species with the apolipoprotein E(APOE) gene and prevalent Alzheimer's disease (AD) (A), the improvement of the power of the associations by meta‐analysis combining Australian Imaging, Biomarkers and Lifestyle (AIBL) and Alzheimer's Disease Neuroimaging Initiative (ADNI) (B), and the causality inference of APOE genotypes to prevalent AD through lipid species by mediation analysis (C). (A) For each participant, we utilized available samples at their last acquired time point (n = 1087) to maximize the number of participants in the association studies. Lipid association studies with APOE were performed using only the control (CN) subset, whereas associations with AD prevalence was examined between the control and AD subsets. Covariates fitted into the models included age, sex, and BMI. Models for ADNI included fasting status, whereas models for AIBL included sample time point. To identify associations that were independent of lipoprotein metabolism, a second set of analyses was performed with further adjustment for clinical lipids (total cholesterol, HDL‐C, and triglycerides). Associations were corrected for multiple comparisons using the method of Benjamini Hochberg (BH). 60 In the Busselton Health Study (BHS), interaction of APOE genotypes and age was examined using a binary cut‐off of 60 years (Table S6). The “clinical lipids” means the linear regression was performed separately with/without clinical lipids adjustment. (B) Associations between APOE genotypes and lipid species and the associations between AD prevalence and lipid species were analyzed using a fixed‐effect inverse‐variance weighted meta‐analysis. Heterogeneity between AIBL and ADNI was assessed using Cochran Q. (C) The mediation analysis was performed on the combined AIBL and ADNI data sets, treating AD as the outcome and APOE genotypes as the treatment. There were two types of mediators: (1) individual lipid species (that showed concordant associations with APOE and AD from the previous analyze) and (2) APOE lipid scores. The lipid scores were created by ridge regression using either the lipid species concordant in association with AD/APOE or all the lipid species to predict APOE ε2/ε4. The models were trained on either the BHS cohort (n = 4384) or the combined AIBL and ADNI cohorts (Control; n = 900), followed with an external validation on the whole population of combined AIBL and ADNI cohorts (n = 1597). The resulting predicted values on the validate set were the APOE lipid scores that were treated as mediators for the mediation analysis
