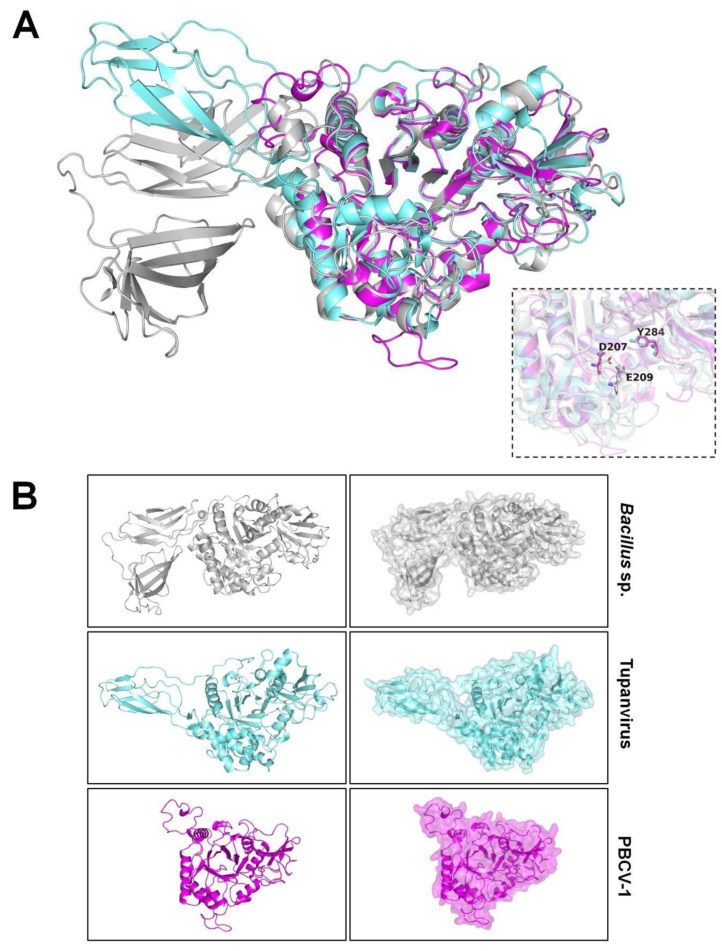
Figure 4.
Models of chitinases from microbial sources. Modeled structures were obtained by SWISS-MODEL (https://swissmodel.expasy.org/ (accessed on 1 October 2022)), represented by drawing. (A) Three-dimensional alignment of chitinases from Bacillus cereus/Bacteria (gray), Tupanvirus/Mimiviridae (cyan), and Paramecium bursaria chlorella virus 1/Phycodnaviridae (pink). The alignment shows an equivalent core for all structures, with an evident difference in size, especially from B. cereus, with two-sided protein fragments, one being correspondent to Tupanvirus and absent in PBCV-1. The box in the right part of the image evidences key common residues at the active site of the enzymes (D207, E209, Y284); (B) Models from each microorganism represented as drawing and surface structures. The modeling of each target sequence was assessed with the best hit available (scored) after employing the search template and build model tools. Model alignment and analysis were assessed with the PyMOL software (v0.99c).