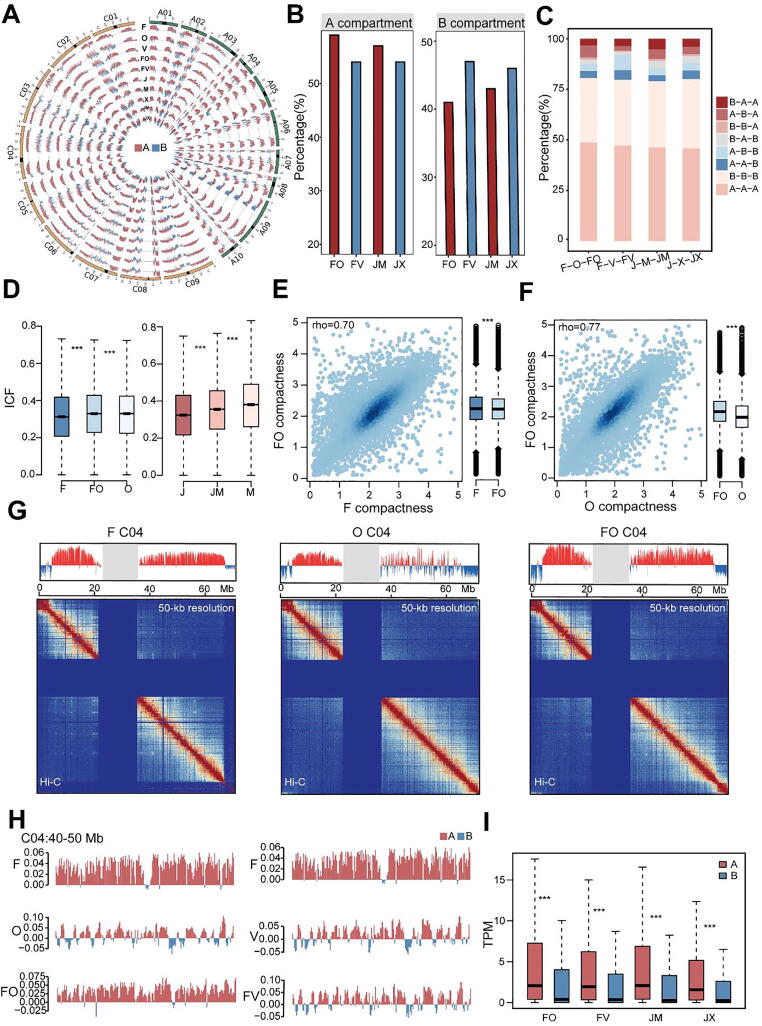
Fig. 2.
Comparison of dynamic changes in the 3D structure of chromatin between parents and hybrids. A, Distribution of compartments in superior and inferior hybrid sets. Red and blue histograms represent A and B compartments, respectively. B, Percentages of different compartments in superior and inferior hybrids. C, Percentages of compartment activity during B. napus hybridization. The order of compartment activity is paternal-maternal-F1. D, Distribution of chromatin interactions in the whole genome of the F-O-FO and J-M−JM groups. E and F, Relationship of chromatin compactness between parents and hybrids between FO and F (E) and FO and O (F). rho, Spearman’s rank correlation coefficient. G, Chromatin interactions in B. napus represented by the C04 chromosome of F-O-FO. The upper track shows the distribution of chromatin compartments in the 3D structure. Red histograms represent A compartments; blue histograms represent B compartments; gray columns indicate masked regions represented as centromere and pericentromeric regions. The lower track shows chromatin interactions at 50-kb resolution. In each heat map, strong interactions are indicated in red and weak interactions in blue, and the dark blue area in the middle represents the pericentromeric regions. H, Features of compartments represented by the first eigenvector from principal component analysis (PCA). Red and blue histograms represent A and B compartments, respectively. I, Dynamic activities of compartments contribute to gene expression in F1 hybrids. The expression level of each gene was normalized to transcripts per million (TPM). *P < 0.05; **P < 0.01; ***P < 0.001 (Wilcoxon rank-sum test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)