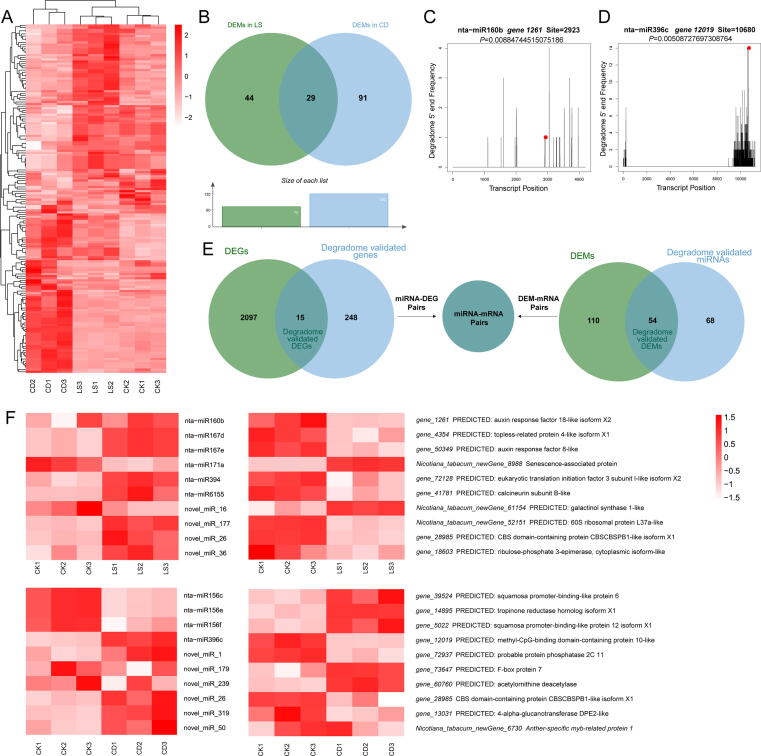
Fig. 3.
Small RNA, degradome sequencing and coherent miRNA-mRNA pairs searching in tobacco seeds. (A) Heat map of enriched miRNAs (the ratio of total; fold change > 1.5; P < 0.05) in three groups. Each group contained three biologically independent replicates (n = 3) used for the heat map. The heat map was generated using normalized (separate z-score computed per gene) expression values in units of TPM. (B) Statistics of DEM number. (C) nta-miR160b-NtARF18 alignments validated by degradome sequencing. (D) nta-miR396c-NtMBD10 alignments validated by degradome sequencing. The red dots represent the cleavage nucleotide positions on the target genes. (E) coherent miRNA-mRNA pairs searching using venn analysis. Degradome validated miRNAs and DEMs were gathered to find degradome validated DEMs, which were then used to find their target genes; Degradome validated genes and DEGs were gathered to find degradome validated DEGs, which were then used to find miRNAs targeting DEGs. (F) A combined view of the expression level of coherent pairs of differentially expressed miRNAs and their target genes. Heat map of coherent miRNAs and genes in two groups. Each group contained three biologically independent replicates (n = 3) used for the heat map. The heat map was generated using normalized (separate z-score computed per gene) expression values in units of TPM and FPKM respectively in miRNAs and genes. The abscissa is the sample name, and the ordinate is the miRNA or gene name with its NR annotation.