Figure 2.
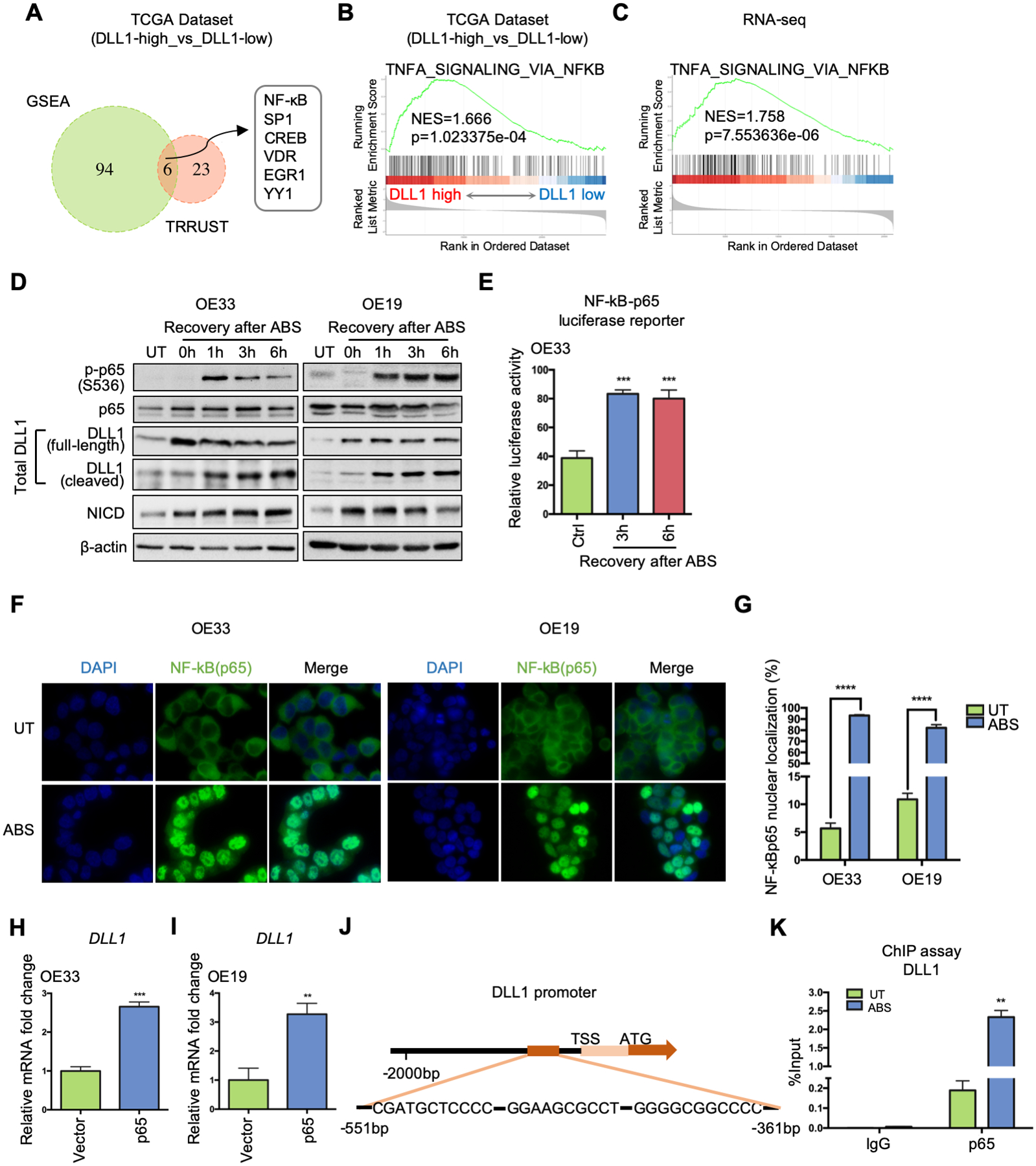
NF-κB is critical for ABS-induced DLL1 transcription by directly binding to DLL1 promoter region. (A) DLL1-expression-based transcription factor enrichment analysis by GSEA and TRRUST website indicates NF-κB as one of the key transcriptional regulators in DLL1-high tumor samples in the TCGA-EAC database. (B) GSEA of NF-κB signaling in the TCGA-EAC database, comparing DLL1-high EAC with DLL1-low EAC. (C) GSEA of NF-κB pathway in the RNA-seq datasets of OE33 cells, comparing control cells with ABS-treated cells (shCtrl vs shCtrl+ABS). (D) Western blots show protein level change of phosphor-p65, total p65, total DLL1 (including full-length ligand and its cleaved fragment), NICD and β-actin during 6h-recovery after ABS exposure in OE33 (left) and OE19 (right) cells. (E) NF-κB-p65 luciferase assay shows NF-κB transcriptional activity were elevated after ABS exposure in OE33 cells. Untreated cells worked as control (Ctrl). (F) Representative immunofluorescent staining images of NF-κB (p65) in OE33 (left) and OE19 cells (right) showing nuclear translocation after ABS exposure. DAPI was used for nuclear staining. (G) Percentage of NF-κB (p65) nuclear localization was quantified using three independent fields. (H) and (I) DLL1 mRNA expression was detected after OE33 (H) and OE19 (I) cells were transfected with a p65 overexpression vector or control (Vector). (J) The ChIP primers amplified the DNA fragment containing three adjacent potential NF-κB binding sites in DLL1 promoter, −551bp to −361bp from transcription start site (TSS). (K) ChIP-qPCR was performed in OE33 cells by using NF-κB antibody to pull down chromatin after ABS exposure, compared to the untreated control. IgG works as control. Statistical data are shown as mean ± SEM. **p<0.01, ***p<0.001 and ****p<0.0001 as calculated by t test for two group comparisons.
