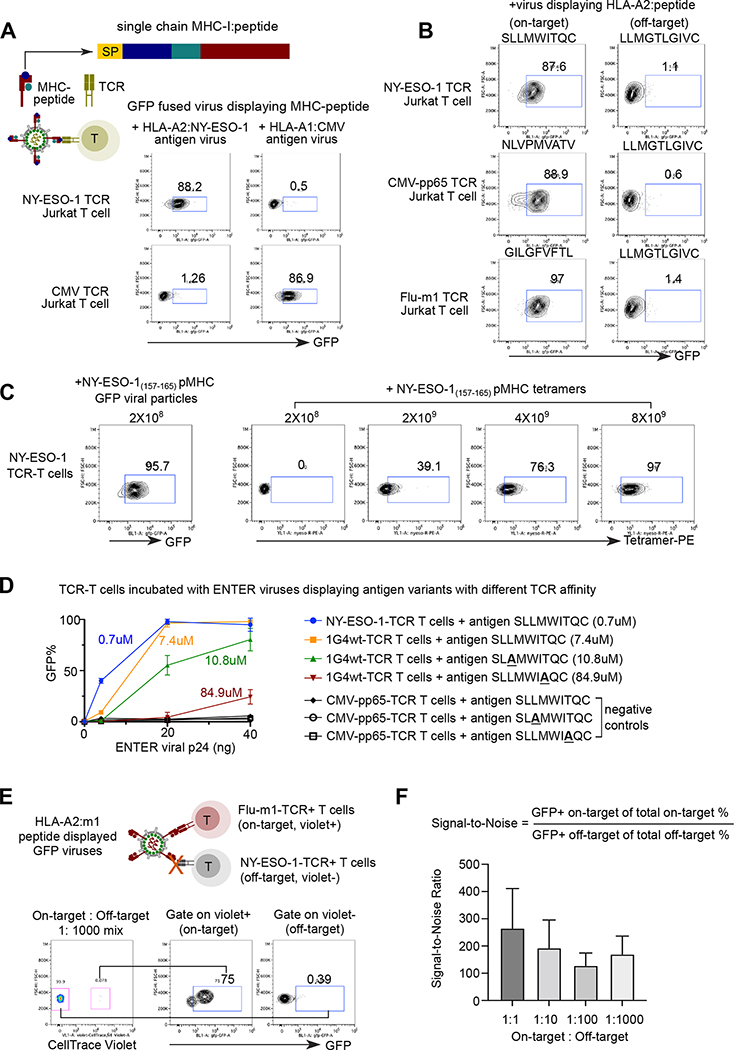
Figure 2. ENTER decodes interaction between pMHC with TCR.
A. Schematic view of pMHC displayed viruses and representative flow plot from two replicates showing GFP signal in specific TCR expressing Jurkat T cells upon incubation of pMHC displayed viruses. GFP fused viruses displaying either a 9-mer peptide (NY-ESO-1157–165) presented by HLA-A0201 allele or an 11-mer CMV peptide (pp65363–373) presented by HLA-A0101 allele were incubated with T cells expressing specific TCRs (e.g., NY-ESO-1157–165-TCR or CMV-pp65363–373-TCR) that recognize the cognate antigens. SP: signal peptide; Peptide: antigen peptide; B2M: Beta-2-Microglobulin.
B. Representative flow plot from two replicates showing GFP signal in Jurkat T cells expressing specific TCRs (e.g., NY-ESO-1157–165-TCR, CMV-pp65495–503-TCR, or Flu-m158–66-TCR) upon incubation of viruses displaying various HLA-A2 presented peptides. HPV16 E782–91 peptide displayed viruses serve as a negative control.
C. Representative flow plot from two replicates showing NY-ESO-1 TCR-T cells upon incubation of 2×108 ENTER viral particles displaying NY-ESO-1157–166 antigen (left) or with different amount of NY-ESO-1157–166 pMHC tetramers.
D. Comparison of binding efficiency (GFP+%) of viruses displaying antigen variants with different TCR affinity. 1G4wt-TCR T cells were incubated with ENTER displaying wild-type of mutant NY-ESO-1157–166 antigen variants for 2 hours. CMV-pp65495–503-TCR T cells were used as negative control. The viral titers were normalized using the p24 protein level.
E. Schematic view of experimental set up (top) and flow cytometry analysis (bottom) of TCR-T cell mixing experiment. Violet dye labeled Flu-m158–66-TCR T cells were mixed with CMV-pp65495–503-TCR T cells at different ratio, and then incubated with HLA-A2:m1 displayed GFP viruses for 2h followed by flow cytometry. Representative flow plot showing 1:1000 mixing of two T cell population and GFP signal of T cells gated on Violet+ and Violet− population.
F. Bar plot showing the signal-to-noise ratio of ENTER in Figure 2E and Figure S2D.
Data are represented as mean +/− SEM in D and F from triplicates.