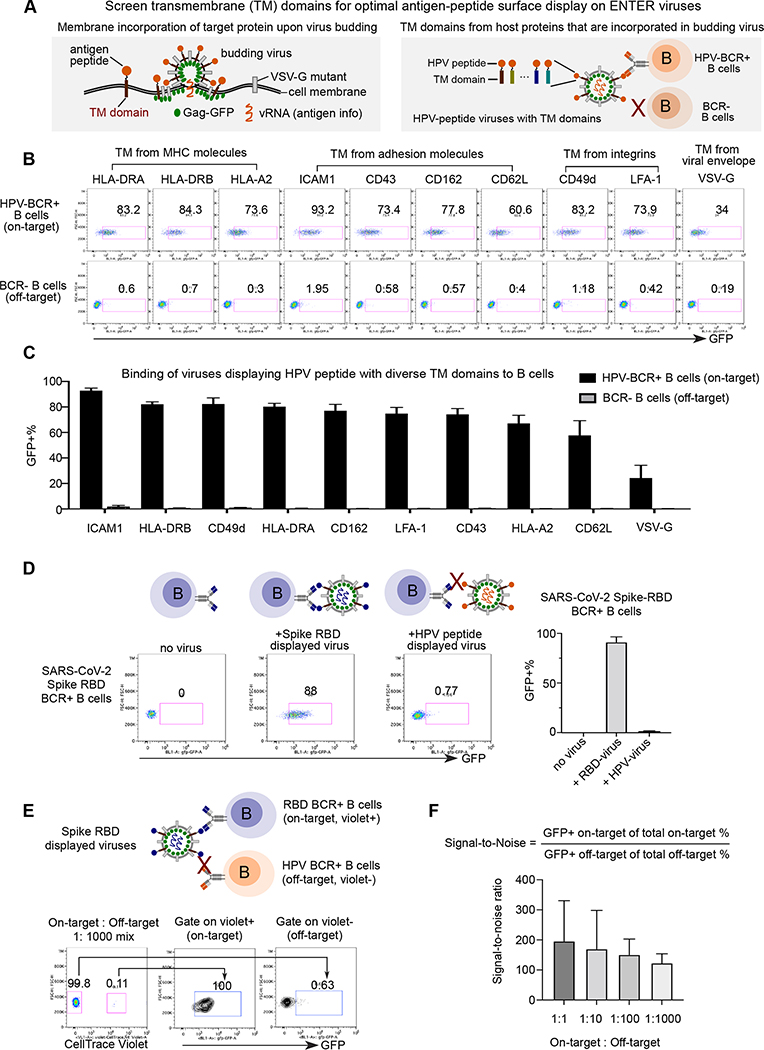
Figure 3. Optimization of ENTER to present intracellular antigens on viral surface and decode interaction between BCR with antigens.
A. Schematic view of experimental design. During the assembly and budding of lentiviruses, certain host cell surface proteins can be incorporated into the surface of viruses. TM domains of host proteins selected from literature are fused with a B cell epitope derived from intracellular antigen HPV16 L2.
B. Representative flow plot from triplicates showing GFP signal in HPV-BCR+ B cells incubated with GFP viruses displaying HPV epitope fused with different TM domains. B cells without BCR expression serve as a negative control.
C. Bar plots showing the percentage of GFP+ B cells from Figure 3B.
D. Representative flow plot from triplicates showing GFP signal in RBD-BCR+ B cells upon incubation of ENTER viruses displaying SARS-CoV-2 Spike RBD antigen or HPV L2 antigen as a negative control (left). Bar plot showing the frequency of GFP+ cells upon incubation of on-target or off-target ENTER viruses (right).
E. Schematic view of experimental set up (top) and flow cytometry analysis (bottom) of B cell mixing experiment. Violet dye labeled RBD-BCR+ B cells were mixed with HPV-BCR+ B cells at different ratio, and then incubated with GFP viruses displaying RBD antigen fused with ICAM1 TM domain. Representative flow plot showing 1:1000 mixing of two B cell population and GFP signal of B cells gated on violet+ and violet− population.
F. Bar plot showing the signal-to-noise ratio of ENTER from Figure 3E and Figure S3C.
Data are represented as mean +/− SEM in C, D, and F from triplicates.