Abstract
Volatile organic compounds (VOCs) have the characteristics of long distance propagation, low concentration, perception, and indirect contact between organisms. In this experiment, Lysinibacillus macroides Xi9 was isolated from cassava residue, and the VOCs produced by this strain were analyzed by the SPME–GC–MS method, mainly including alcohols, esters, and alkanes. By inoculation of L. macroides Xi9, VOCs can promote the growth and change the root-system architecture of Arabidopsis seedlings. The results showed that the number of lateral roots, root density, and fresh weight of Arabidopsis seedlings were significantly higher (p ≤ 0.01), and the number of roots hair was also increased after exposure to strain Xi9. Compared with the control group, the transcriptome analysis of Arabidopsis seedlings treated with strain Xi9 for 5 days revealed a total of 508 genes differentially expressed (p < 0.05). After Gene Ontology enrichment analysis, it was found that genes encoding nitrate transport and assimilation, and the lateral root-related gene ANR1 were up-regulated. The content of NO3− and amino acid in Arabidopsis seedlings were significantly higher from control group (p ≤ 0.01). Plant cell wall-related EXPA family genes and pectin lyase gene were up-regulated, resulting cell elongation of leaf. SAUR41 and up-regulation of its subfamily members, as well as the down-regulation of auxin efflux carrier protein PILS5 and auxin response factor 20 (ARF20) led to the accumulation of auxin. These results indicated that VOCs of strain Xi9 promote Arabidopsis seedlings growth and development by promoting nitrogen uptake, regulating auxin synthesis, and improving cell wall modification.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-022-01268-3.
Keywords: Lysinibacillus macroides, Transcriptome, VOCs, Root-system architecture, Auxin, Cell wall modification
Introduction
Plant growth-promoting bacteria (PGPB) can benefit plant growth via quite a few of mechanisms and extensively exist in the rhizosphere of many plants (Mishra et al. 2021; Qi et al. 2018), and they are environment-friendly and sustainable resource (Ferreira et al. 2019). The mechanisms of PGPB promoting plant growth include the production of plant hormones such as auxin and cytokinin, the increase of available minerals (phosphorus, iron and nitrogen), the inhibition of pathogens of soil-borne plant diseases and the induction of plant systemic resistance (Batista et al. 2021; Majeed et al. 2018; Mishra et al. 2021). Lysinibacillus macroides was isolated from cow dung for the first time in 1947, which is a spore-forming, Gram-positive bacterium (Coorevits et al. 2012; Liu et al. 2015). Lysinibacillus macroides could inhibit biofilm and reduce colonization of Salmonella (a human pathogen). As a biocontrol agent, L. macroides could reduce the amount of Salmonella in plants (Karmakar et al. 2022). However, the growth-promoting trait of L. macroides has not been reported until now.
Volatile organic compounds (VOCs) have shown to possess similar abilities to PGPB (Tahir et al. 2017a), including the promotion of plant growth (Syed-Ab-Rahman et al. 2019), induction of plant system resistance (Ryu et al. 2004), accumulation of plant secondary metabolites, inhibition of plant pathogens (Chen et al. 2021; Santoro et al. 2011), and enhancement of plant salt tolerance (Li et al. 2021). Microbial volatile compounds have characteristics of low molecular weight, weak polarity, high vapor pressure, low boiling point and lipophilicity (Chen et al. 2021; Schöller et al. 2002; Weisskopf et al. 2021). Compared with other secondary metabolites of microorganisms, microbial VOCs have the advantage of being able to travel long distances, mediating indirect contact between organisms, and low concentrations can be perceived. Therefore, VOCs have shown another mechanism of promoting plant growth.
Ryu et al. (2003) firstly found that 3-hydroxy-2-butanone and (2R,3R)-butanediol generated by two Bacillus could stimulate growth and cause systemic resistance of Arabidopsis, increase the production of auxin in Arabidopsis root, up-regulate the expression of the auxin transporter gene, and promote cell enlargement (Ryu et al. 2004; Zhang et al. 2007). Park et al. (2015) reported that 13-Tetradecadien-1-ol and other 10 compounds generated by Pseudomonas fluorescens SS101 could facilitate tobacco growth and development. Burkholderia gladioli BBB-01 generates 2,5-dimethylfuran which could control plant fungal disease (Lin et al. 2021). Moreover, Tahir et al. (2017b) demonstrated that albutamol and 1,3-propanediole generated by Bacillus subtilis SYST2 could promote tomato growth, with a significantly increased photosynthesis rate and stomatal conductance after inoculation with VOCs compared with the control. Nearly 1000 species of bacteria have reportedly generated approximately 2000 different VOCs (Lemfack et al. 2018). Analysis of microbial VOCs has a major impact on biotechnology, medicine, and agricultural applications and basic science.
Although a large number of researches have demonstrated that microbial VOCs can mediate plant development, the molecular and genetic basis of research is extensive and little is known about the overall transcriptional activity of plants. Through high-throughput sequencing, transcriptomics provides comprehensive and rapid access to almost all transcripts of a particular organ or tissue in a given state. In this experiment, the VOCs from L. macroides Xi9 promoted the growth of Arabidopsis seedlings and changed the root-system architecture. The growth-promoting mechanism of VOCs generated by strain Xi9 was further revealed by transcriptome sequencing. This study provides a theoretical basis for strain L. macroides Xi9 as a microbial fertilizer.
Materials and methods
Bacterial strain and culture condition
Lysinibacillus macroides Xi9 isolated from cassava residue was cultivated on Luria–Bertani (LB) agar medium and cultivated at 37 °C. The bacteria was stored in liquid LB medium containing 25% glycerol and maintained at − 20 °C for a long-term storage.
Phylogenetic analysis and identification of the strain
Total DNA of strain Xi9 was extracted using the Bacteria Genomic DNA kit (CWBIO, Beijing, China) according to the manufacturer’s protocol. The specialized primers used for PCR amplification were 27 forward (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492 reverse (5′-GGTTACCTTGTTACGACTT-3′). The PCR products were sequenced by RuiBiotech. The DNA sequences were compared with GenBank using the National Center for Biotechnology Information (NCBI) BLAST program (http://www.ncbi.nlm.nih.gov/blast). Based on Jukes-Cantor model and Neighbor-joining method, the phylogenetic trees were constructed with the MEGA 5.0 program (Cao et al. 2021).
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) seedlings were used because of their suitability for in vitro growth. The seeds were washed with 70% ethanol for 5 min and 2.6% bleach for 10 min to surface-sterilize, washed with sterile water for five times, and then placed on 1.5 mL centrifuge tube to vernalized for 2 days at 4 °C. The seeds were cultured on 9 cm petri dishes containing 1/2 Murashige and Skoog salt (MS) medium (Hopebio, Qingdao, China), 0.8% agar and 2% sucrose, at pH 5.7. These petri dishes were placed in plant light incubator with a photoperiod of 8 h of darkness and 16 h of light. Petri dishes were tilted at a 65-degree angle to facilitate roots grow on the surface of the medium.
Qualitative analysis of indole-3-acetic acid of strain Xi9 production
Strain Xi9 was inoculated into LB liquid medium containing 200 mg L-tryptophan and cultivated at 28 °C, 180 rpm for 4 days. The bacterial culture suspension was centrifuged at 10,000 rpm for 10 min, and then the Salkowski reagent (0.5 M FeCl3 1 mL, 35% HClO4 50 mL) was added with an equal volume of the supernatant for color development for 30 min in the darkness (Patten and Glick 2002; Khan et al. 2014). The observed red color means the strain could produce indole-3-acetic acid.
Inoculation experiments
Strain Xi9 was cultured on LB agar medium as described above for 12 h, inoculated into liquid LB medium to the exponential phase, and then followed by dilution to 1 × 108 cfu/mL. Plastic petri dishes (9 cm) containing a center partition were prepared, with one side containing 1/2 MS medium and the other side containing LB medium. Arabidopsis seedlings that germinated for four days were transferred to 1/2 MS medium (5 seedlings per plate). To investigate the VOCs effect of strain Xi9 on Arabidopsis seedling, two groups were set up: (i) 10 μL of strain Xi9 were added to the other side of the plate containing LB medium (Xi9); (ii) 10 μL of liquid LB medium were added to the other side of the plate containing LB medium (control). These plate were sealed by the parafilm, placed in the plant light incubator at random and tilted at a 65-degree angle to facilitate roots growth. Thus, the VOCs from strain Xi9 could directly interact with plants. Each group was designed with three biological replicates. Seven days after inoculation, the fresh weight, primary root length, and density of lateral roots of the Arabidopsis seedling were measured (Tahir et al. 2017b).
Determining the sampling time for Illumina sequencing by quantitative real-time PCR (qRT-PCR)
Total RNAs were obtained from plants were exposed to VOCs of strain Xi9 for 3 days, 5 days, and 7 days using TRIzol reagent (CWBIO, Beijing, China) according to the manufacturer’s instruction. The cDNA synthesis using EasyScript® All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (Transgene Biotech, Beijing, China). qRT-PCR was carried out using SYBR® Premix Ex Taq polymerase (Transgene Biotech, Beijing, China) with the Rotor Gene Q Real-time PCR System (Qiagen, Germany). The program started with a pre-denaturation at 94 °C for 30 s, then 94 °C for 5 s, 60 °C for 15 s, and 72 °C for 20 s for 40 cycles, subsequently extended at 72 °C for 5 min. Every group and each sample was independently assessed in triplicate. The 2−ΔΔCt method was used to analyze the data. Six genes were chosen to determine the appropriate time for Illumina sequencing: two genes related to auxin (ARF20 encodes auxin response factor 20, IAA5 encodes indole-3-acetic acid inducible 5), two genes related to nitrate transporter (NRT1.8 encodes nitrate transporter, NRT2.1 encodes high-affinity nitrate transporter), one gene related to cell wall modification (EXPA17 encodes expansin 17), and one gene related to ABC transporter (ABCB15 encodes ABC transporter family protein). The primers used for qRT-PCR are shown in Table S1.
Illumina sequencing
For transcriptome analysis, we conducted three independent experiments to obtain plant materials, and each corresponding to one biological replicate. Using mirVana miRNA Isolation Kit (Ambion, USA) to extract total RNA from Arabidopsis seedlings according to the manufacturer’s instruction. The Agilent 2100 Bioanalyzer (Agilent Technologies, USA) was applied to detect RNA integrity. Samples were subject to subsequent analysis with an RNA Integrity Number (RIN) ≥ 7. Qualified total RNA was further purified by an Agencourt AMPure XP Kit (Beckman Coulter, Brea, CA, USA). After total RNA purified, magnetic beads with Oligo (dT) were used to enrich mRNA; the mRNA were disrupted into approximately 200-nt fragments after adding disruption reagent. The cDNA fragments were purified using AMPure XP beads. The cDNA libraries were set up by the TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer’s protocol. Based on the Illumina sequencing platform (HiSeqTM X Ten), these six libraries were sequenced, and 125 bp/150 bp paired-end reads were produced. All samples were sequenced on OE Biotech.
Differentially expressed genes (DEGs) identification and functional annotation
FPKM (fragments per kilobase of exon per million fragments mapped) (Trapnell et al. 2010) was computed by htseq-count and cufflinks to obtain the read counts for each gene (Anders et al. 2015). The DESeq R package functions estimate Size Factors and nbinom Test were separately used to identify DEGs (Anders and Huber 2012). For significantly differential expression, a p < 0.05 and |log2FC|> 1 were used as the threshold. Based on the hypergeometric distribution, Gene ontology (GO, http://www.geneontology.org/) enrichment analysis of DEGs was carried out using R.
Collection and analysis of bacterial volatiles
Strain Xi9 were inoculated into 20 mL of LB sealed with 4 layers of foil paper and inoculated at 37 °C under agitation at 180 rpm for 2 days to accumulate VOCs. The LB medium as a control group (CK). VOCs were extracted through a blue SPME fiber (Polydimethylsiloxane/Divinylbenzene, PDMS/DVB) (Supelco, Inc., Bellafonte, PA, U.S.A.). The SPME fiber tip was exposed to the upper air of the sample. The sample was stirred at medium speed with a magnetic stirrer, heated to 40 °C, and adsorbed and extracted for 30 min (Lin et al. 2021). The needle was inserted into the gasification chamber of the gas chromatograph. The fiber tip was desorbed at 230 °C for 3 min. The samples were detected by a GC–MS-QP2010 from Shimadzu (70 eV; Kyoto, Japan) with a Rtx-5 column (30 m × 0.25 mm × 0.25 mm; J&WScientiWc, Folsom, CA, USA). The operating conditions included He (1 mL/min) as the carrier gas. The primary oven temperature was 40 °C, which was maintained for 3 min. The oven was then heated to 95 °C at the rate of 10 °C/min, further turned to 230 °C at the rate of 3 °C/min for 5 min (Gutiérrez-Luna et al. 2010). The VOCs were recognized by comparing with mass spectra from the library (NIST14 and NIST14s).
IAA content analyses of plants
Samples were collected after 7 days of treatment. The content of IAA was determined by HPLC (Kelen et al. 2004; Pérez-Pastrana et al. 2021). Fresh samples (0.15 g) were ground in 1 mL precooled 20% methanol, and extracted overnight at 4 °C. Samples were centrifuged at 10,000 rpm for 10 min, and the extracts were stored. Evaporating under reduced pressure at 40 °C to eliminate the organic phase of the extracts (approximately 0.3 mL solution). A total of 0.5 mL petroleum ether was added for extraction and decoloration. 1 mol/L citric acid solution was added to samples to adjust the pH to 2–3, followed by two extractions with 1 mL ethyl acetate, blew dry with nitrogen. The extracts were then dissolved, mixed, filtered and loaded onto HPLC (Kromasil) C-18 column (250 mm × 4.60 mm, 5 μm).
Nitrate and amino acids analyses
The contents of nitrate (NO3−) and amino acids were detected using 7-day-old plants were treated or not with strain Xi9 (1 × 108 cfu/mL). Using the salicylic acid method to measure NO3− content (Xu et al. 2016). Briefly, the material was put into a 1.5 mL centrifuge tube, weighed and subsequently milled to powder in liquid nitrogen. Then, adding 1 mL of deionized water to the tube, and boiling the mixture at 100 °C for 15 min, followed by centrifuging at 13,000 rpm for 10 min, and the supernatant was used as the sample solution for the assay. 100 μL of sample solution and 400 μL of 5% (W/V) salicylic acid-sulfuric acid solution were mixed and reacted for 20 min at room temperature. The mixture was followed by adding 9.5 mL of 8% (W/V) NaOH solution, mixed, and cooled to room temperature. We used 0.1 mL deionized water as a blank reference and quantified the optical density at the wavelength of 410 nm. The content of nitrate was calculated following the formula: Y = C·V/W (C, concentration of nitrate obtained based on the OD410 in a regression equation; V, the total capacity of extracted samples; W, sample weight; Y, nitrate content). A standard curve was prepared with KNO3 at a concentration of 10–120 mg/L, and the regression equation was obtained from the standard curve.
The total amount of free amino acids in plants was measured using the ninhydrin method (Sun et al. 2006). Arabidopsis seedlings (0.5 g) were put into 5 mL of 10% acetic acid solution, subsequently centrifuged at 10,000 rpm for 5 min. Then, 500 μL supernatant was brought to 25 mL using 0.2 M acetate buffer, pH 5.4. The sample extract (10 mL) was boiled at 100 °C for 15 min. Next, 1 mL sample solution, 1 mL 0.2 M acetate buffer, 100 μL 0.3% ascorbic acid solution, and 3 mL ninhydrin solution were mixed, boiled at 100 °C for 15 min, and diluted to 20 mL. The blank reference consisted of 1 mL of deionized water, and the absorbance was measured at a wavelength of 580 nm. The total amount of free amino acids was determined using the formula: Y = C·V1/W·V2 (Y, the total amount of free amino acids; C, amino acid concentration calculated based on the OD580 in a regression equation; V1, dilution total volume; W, sample weight; V2, colorimetric total volume). A standard curve was prepared with leucine at a concentration of 1–5 μg/mL, and a regression equation was derived from the standard curve.
Statistical analysis
The results were analyzed using the statistical software SPSS version 19.0 (SPSS Inc., Chicago, IL, USA) by analysis of variance (ANOVA) and Duncan’s multiple-range test (p < 0.05). The significance was determined by Duncan’s (D) test (Wang et al. 2020).
Results
General characteristics of strain Xi9
Strain Xi9 was isolated by our group from cassava residue, which was identified based on 16S rRNA gene sequence analysis as L. macroides (formerly named Lineola longa and Bacillus macroides) (Fig. 1) (Coorevits et al. 2012), and it was preserved in the China General Microbiological Culture Collection Center (CGMCC) under No.15583. After incubation of strain Xi9 on LB medium for 24 h at 37 °C, the colonies were moist, round, and cream-colored, with smooth edges and surfaces, and have diameters of 3–5 mm. The morphological characteristics of the bacteria were rod-shaped, single-cell. The strain Xi9 was stained as Gram-positive strain and could form spores. Oxidase production was negative and catalase production was positive for this strain. Aesculin, starch, and gelatin were not hydrolyzed by this strain. No acid was produced from this strain and the Voges–Proskauer assay was positive. Strain Xi9 was verified to produce indole-3-acetic acid (Fig. S1).
Fig. 1.
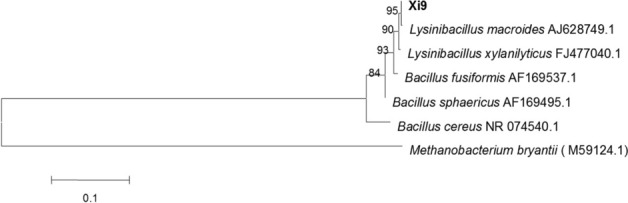
Phylogenetic trees of the strain Xi9 based on 16S rRNA gene sequence. The phylogenetic trees were built using the MEGA 5.0 program, displaying the phylogenetic location of strain Xi9 and some other related species based on the sequence of 16S rRNA gene. Bootstrap values (> 50%) are represented at the branch points based on 1000 replications. Bar, 0.1 substitutions per nucleotide position
Phenotypic and physiological features of Arabidopsis seedling are affected by L. macroides Xi9
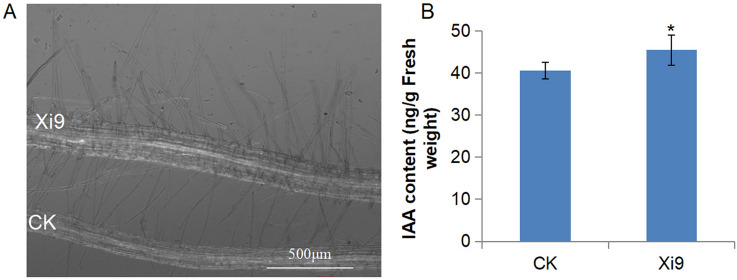
After treatment of the strain Xi9 for 7 days, the phenotype of Arabidopsis seedlings of the Xi9 group showed much stronger expression than control group (Fig. 2A). In the Xi9 group, the number of lateral roots, root density, and fresh weight of Arabidopsis seedlings reached a considerable difference (p ≤ 0.01), compared with the control group, whose values were 108.6%, 148.63%, and 137.5% higher, respectively (Fig. 2B–D). However, the primary root length did not increase (Fig. 2E). Under the microscope, it was observed that the primary root became thicker in the Xi9 group (Fig. 2F, S2). The VOCs released by strain Xi9 could influence the root and shoot development and growth in Arabidopsis seedlings.
Fig. 2.
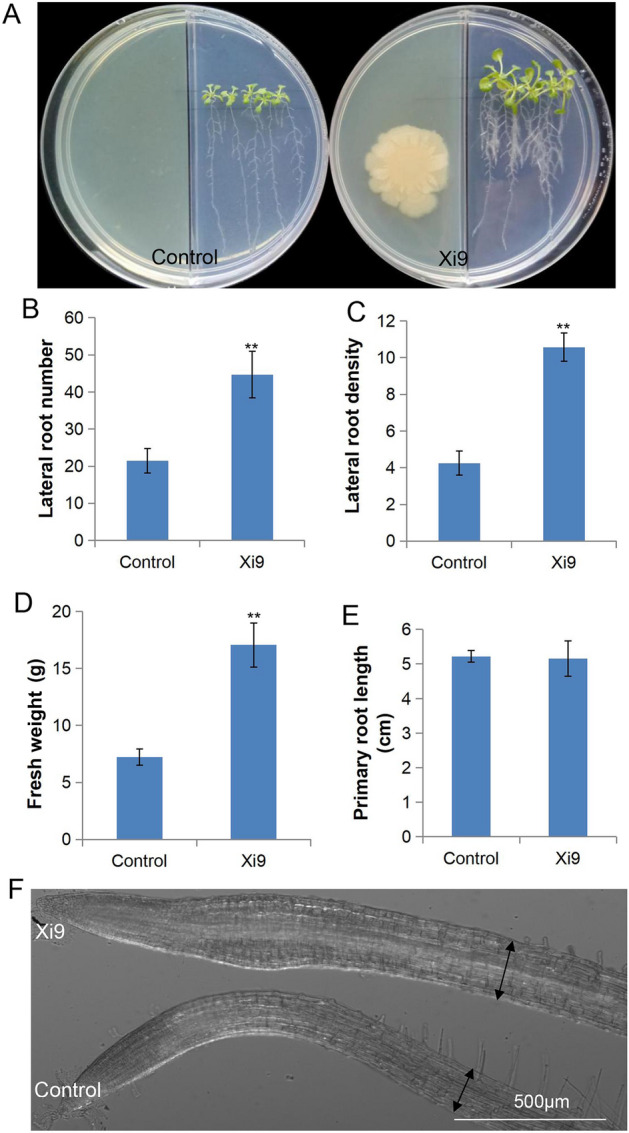
Growth condition of Arabidopsis seedlings after inoculation by L. macroides Xi9. Four-day-old Arabidopsis seedlings growing in Petri dishes tilted 65 degrees, inoculated with L. macroides Xi9 or liquid LB medium. After 7 days of treatment, A the seedling phenotype, B lateral root numbers, C root density, D fresh weight, E primary root length, and (F) primary root phenotype determined. **indicates p ≤ 0.01
The volatile emission by L. macroides Xi9
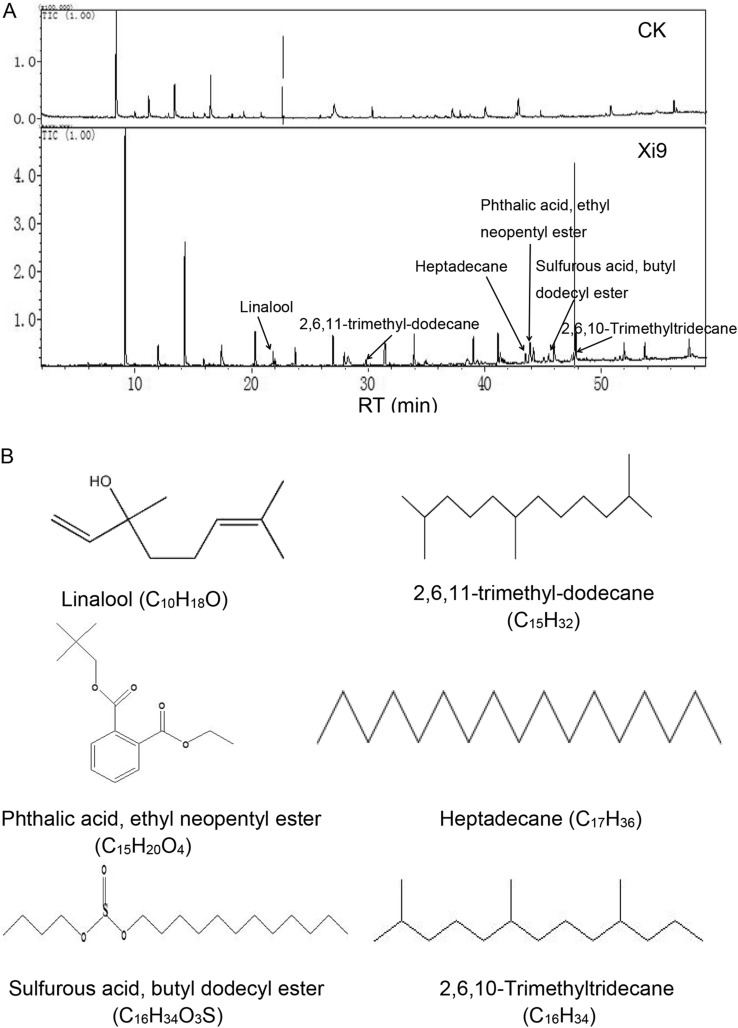
We detected the component of VOCs generated by strain Xi9 cultured in 20 mL liquid LB medium for 2 days at 37 °C and measured the compound by SPME–GC–MS. Six volatile compounds with relatively high peak areas were identified including three alkanes (2,6,11-trimethyl-dodecane, 2,6,10-trimethyltridecane and heptadecane), two esters (phthalic acid, ethyl neopentyl ester, and sulfurous acid, butyl dodecyl ester), and linalool (Table 1, Fig. 3A, B).
Table 1.
GC–MS profile of VOCs generated by L. macroides Xi9
| RT (min) | Total area (%) | Possible compounds |
|---|---|---|
| 22.011 | 0.41 | Linalool |
| 29.801 | 0.6 | 2,6,11-trimethyl-Dodecane |
| 43.512 | 1.25 | Heptadecane |
| 43.883 | 4.85 | Phthalic acid, ethyl neopentyl ester |
| 45.476 | 0.88 | Sulfurous acid, butyl dodecyl ester |
| 47.668 | 1.24 | 2,6,10-Trimethyltridecane |
Fig. 3.
GC–MS profile of volatiles of L. macroides Xi9. L. macroides Xi9 were inoculated into 20 mL of LB and inoculated at 37 °C and 180 rpm for 2 days to accumulate volatiles. A Chromatographic profiles of volatiles produced by strain Xi9, B structural formulas of VOCs
The appropriate time for Illumina sequencing
The changes of 6 genes (ARF20, EXPA17, IAA5, NRT1.8, NRT2.1, and ABCB15) in Arabidopsis seedlings treated by strain Xi9 for different time points were detected by qRT-PCR. The results showed that the expression of ARF20 and IAA5 reached the highest level at 5 days. The expression of EXPA17 and NRT1.8 showed no significant changes at 5 and 7 days but all higher than at 3 days (p < 0.05), and the expression of NRT2.1 reached the highest level at 7 days (Fig. 4). We finally decided to sequence the sample on day 5.
Fig. 4.
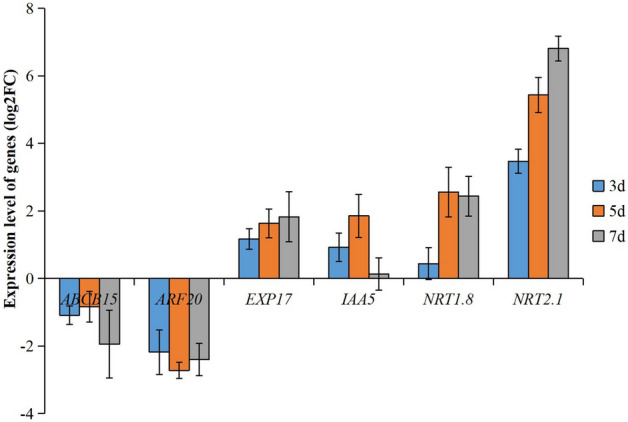
qRT-PCR analysis of the gene expression profile at different time points. qRT-PCR was performed to assess the expression levels of six genes (ARF20, EXPA17, IAA5, NRT1.8, NRT2.1, ABCB15) at 3, 5 and 7 days
Differentially expressed genes (DEGs) of Arabidopsis seedlings affected by VOCs of strain Xi9
To achieve a comprehensive transcriptome level in Arabidopsis seedlings affected by VOCs of strain Xi9, six libraries were constructed from Arabidopsis seedlings (three biological replicates per group). The raw reads produced by six libraries ranged from 47,038,086 to 48,810,854. The scale of clean reads was above 97% for each library after removing the low quality reads (Table S2). For the expression profile of Arabidopsis seedlings affected by VOCs from strain Xi9, the FPKM was used to calculate gene expression. Based on the threshold consisting of a p < 0.05 and |log2FC|> 1, a total of 508 genes were differentially expressed after exposure to VOCs of strain Xi9. 168 genes were up-regulated and 340 genes were down-regulated (Table S3). Highly up-regulated genes including high-affinity nitrate transporter 2.6 (At3g45060), phosphate transporter (At5g43360), high-affinity nitrate transporter 2.1 (At1g08090), and MADS-box transcription factor (At2g14210). Highly down-regulated genes containing 1-deoxy-D-xylulose 5-phosphate synthase 1 (At3g21500), acyl-CoA N-acyltransferases (NAT) superfamily protein (At2g39030), and UDP-glucosyl transferase 76E12 (At3g46660).
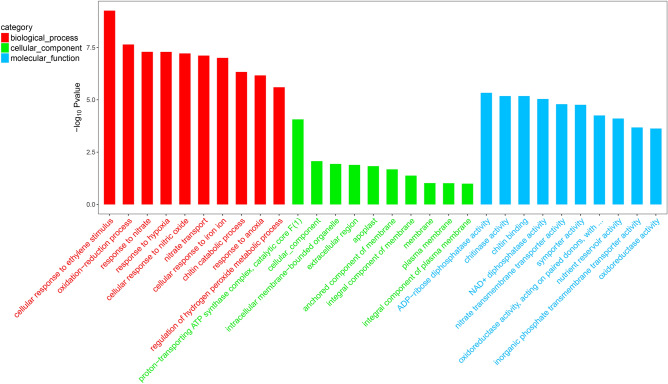
To identify the functions of DEGs of Arabidopsis seedlings, annotated functions of DEGs were analyzed using the GO database. In the GO analysis, the largest category was biological processes among the three major categories (Fig. 5). In the biological process, cellular response to ethylene stimulus (GO0071369), oxidation–reduction process (GO0055114), and response to nitrate (GO0010167) were the three largest groups. In the cell component, the most abundant group was proton-transporting ATP synthase complex, catalytic core F(1) (GO0045261). For the molecular function category, the three most abundant groups were ADP-ribose diphosphatase activity (GO0047631), chitinase activity (GO0004568) and chitin binding (GO0008061). The results preliminarily indicated that VOCs generated by strain Xi9 mainly affected the transcription of genes associated with plant hormone and nitrate metabolism.
Fig. 5.
The GO enrichment analysis of the DEGs of Arabidopsis seedlings. The top 30 GO categories assigned to the DEGs of Arabidopsis seedlings during inoculation with VOCs of L. macroides Xi9
Verification of DEGs using qRT-PCR
To verify the reliability of the RNA-seq data, several genes in Arabidopsis seedlings were detected by qRT-PCR. In Illumina sequencing data, the genes related to nitrate transporter (NRT1.8, NRT2.1, and NRT2.6) in Arabidopsis seedlings were up-regulated by 3.00-, 6.60-, and 10.43-fold, respectively. In the qRT-PCR result, the expression patterns of the genes NRT1.8, NRT2.1, and NRT2.6 were similar with the Illumina sequencing data (Fig. S3). In the qRT-PCR, the genes related to cell wall modification (EXPA12, EXPA17, PILS5, and BG2) were significantly changed in the Xi9 group than control group (Fig. S3), and in Illumina sequencing data of Arabidopsis seedlings, these genes were changed 2.39-, 3.62-, -1.29-, and -3.40-fold, respectively. In the qRT-PCR, the gene ANR1 related to lateral root was up-regulated, and in Illumina sequencing data, the gene was up-regulated 5.65-fold. The expression pattern of these selected genes in qRT-PCR was similar with the Illumina sequencing data, suggesting that the Illumina sequencing data were reliable.
VOCs from strain Xi9 influence the auxin synthesis in Arabidopsis seedlings
Auxin can affect plant growth and development in various aspects by mediating cell expansion and division. In our results, some of genes involved in auxin synthesis were up-regulated, including SAUR41 and its subfamily members (Table 2). In contrast, the gene PILS5, annotated as a auxin efflux carrier family protein involved in the polar transport of auxin, was down-regulated, resulting in the accumulation of auxin in root and an increase in number of root hair (Fig. 6A). The gene ARF20, a negative regulator of the auxin response factor, was down-regulated to promote auxin production. As expected, the IAA content of Arabidopsis seedlings in the Xi9 group was significantly higher than the control group (p < 0.05, Fig. 6B), which demonstrated that VOCs of strain Xi9 could influence auxin synthesis in Arabidopsis seedlings.
Table 2.
Auxin-related genes regulated by VOCs of L. macroides Xi9
| Gene id | Putative function | Gene name | log2FC |
|---|---|---|---|
| At1g16510 | SAUR-like auxin-responsive protein family | SAUR41 | 1.3074 |
| At1g79130 | SAUR-like auxin-responsive protein family | SAUR40 | 2.1000 |
| At1g29490 | SAUR-like auxin-responsive protein family | SAUR68 | 1.8818 |
| At1g76190 | SAUR-like auxin-responsive protein family | SAUR61 | − 1.7438 |
| At2g17500 | Auxin efflux carrier family protein | PILS5 | − 1.2885 |
| At3g28345 | ABC transporter family protein | ABCB15 | − 1.6990 |
| At4g32280 | indole-3-acetic acid inducible 29 | IAA29 | 1.0070 |
| At1g15580 | indole-3-acetic acid inducible 5 | IAA5 | 1.6146 |
| At1g35240 | auxin response factor 20 | ARF20 | − 1.4166 |
Fig. 6.
The root hair and IAA content of Arabidopsis seedlings. A Arabidopsis seedlings were inoculated with L. macroides Xi9 for 7 days, and the root hairs of Arabidopsis seedlings increased. B The IAA content of Arabidopsis seedlings following 7 days of exposure were measured by HPLC. *indicates p < 0.05
Cell wall modification was induced by Xi9 VOCs
In addition, the root hair is derived from a single epidermal cell. It is believed that the cell wall components loosening initiates and facilitates root hair growth. The loosening may be regulated by cell wall-loosening expansin proteins (EXPs). The genes EXPA7, EXPA12, EXPA16, and EXPA17 were all up-regulated after exposure to VOCs of strain Xi9 for 5 days, compared with the control group (Table 3). Some other genes encoding cell wall modification factors important for reducing cell wall rigidity were mainly up-regulated in the Xi9 group, including pectate lyase, aspartic-type endopeptidase, and β-1,3-glucanase. Down-regulation of the BG2 gene (encodes a beta-1,3-glucanase) reduced callose synthesis and cell wall rigidity, increased cell wall elasticity, and promoted cell elongation (Fig. 7A, B). These findings revealed that the EXPs genes promoted the growth and development of root hairs and lateral roots.
Table 3.
Cell wall modification genes that were differentially regulated by VOCs of L. macroides Xi9
| Gene id | Putative function | Gene name | log2FC |
|---|---|---|---|
| At3g15370 | Expansin 12 | EXPA12 | 2.3868 |
| At4g01630 | Expansin A17 | EXPA17 | 3.6152 |
| At3g55500 | Expansin A16 | EXPA16 | 1.8004 |
| At2g43890 | Pectinlyase-like superfamily protein | – | 4.9727 |
| At1g05650 | Pectin lyase-like superfamily protein | – | 2.6535 |
| At5g48430 | Eukaryotic aspartyl protease family protein | – | 2.7162 |
| At3g57260 | Beta-1,3-glucanase 2 | BG2 | -3.4018 |
| At1g62440 | Leucine-rich repeat/extensin 2 | LRX2 | 1.0733 |
| At1g12560 | Expansin A7 | EXPA7 | 0.8419 |
Fig. 7.
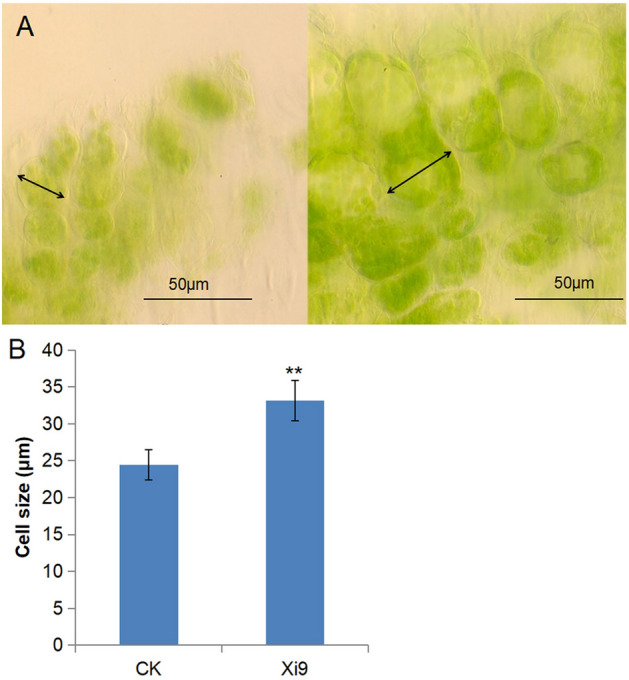
The phenotype of the cell in Arabidopsis seedlings. Arabidopsis seedlings were exposed to VOCs for 7 days, and the cell of Arabidopsis seedlings become elongated compared with that of the control group (A), (B). **indicates p ≤ 0.01
Nitrogen content and lateral roots were influenced by Xi9 VOCs
Nitrate (NO3−) is the best obtainable form of nitrogen in soil. Compared with the control group, after inoculation with strain Xi9 for 5 days, the transcription of NRT2.1, NRT2.3, NRT2.5, NRT2.6, NRT3.1 and NRT1.8 were all up-regulated in Arabidopsis seedling (Table 4). Meanwhile, the contents of nitrate and amino acids of Arabidopsis seedlings in the Xi9 group showed considerably higher expression than control group (p < 0.05) (Fig. 8A, B). The results illustrated that VOCs of L. macroides Xi9 positively influence the content of nitrogen in Arabidopsis seedlings.
Table 4.
Nitrate-related genes regulated by VOCs of L. macroides Xi9
| Gene id | Putative function | Gene name | log2FC |
|---|---|---|---|
| At1g08090 | Nitrate transporter 2.1 | NRT2.1 | 6.6016 |
| At4g21680 | Nitrate transporter 1.8 | NRT1.8 | 3.0024 |
| At3g45060 | High affinity nitrate transporter 2.6 | NRT2.6 | 10.4284 |
| At5g60780 | Nitrate transporter 2.3 | NRT2.3 | 2.0331 |
| At2g14210 | MADS-box transcription factor | ANR1 | 5.6489 |
| At5g50200 | Nitrate transmembrane transporter | NRT3.1 | 1.5398 |
| At4g14358 | High affinity nitrate transporter | NRT2.5 | 2.9014 |
Fig. 8.
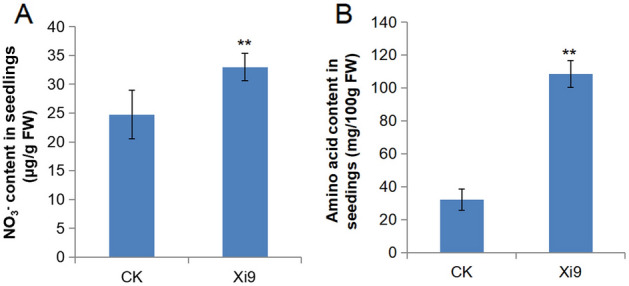
The content of nitrate and amino acid in Arabidopsis seedlings. After Arabidopsis seedlings were exposed to VOCs for 7 days, the contents of nitrate (A) and amino acid (B) in Arabidopsis seedlings were measured. The treated group indicated significant differences from the control group (p ≤ 0.01). **, indicates p ≤ 0.01
In Arabidopsis, the ANR1 MADS-box gene was recognized as an important regulator for the growth of lateral roots during reaction with external nitrate (NO3−) signals (Gan et al. 2012). After Arabidopsis seedlings exposed to VOCs from strain Xi9, the gene ANR1 was up-regulated. Correspondingly, the length and number of lateral roots were considerably greater than control group (Fig. 2A, B).
Discussion
Many researches have reported that PGPB have the ability to synthesize and release VOCs spreading for a long or short distance in the soil (Santoro et al. 2011; Tilocca et al. 2020). Plants can perceive and react to microbial VOCs, which can induce systemic resistance of plant and regulate plant development and growth (Choudhary et al. 2016; Li et al. 2021; Tahir et al. 2017a). In this study, we revealed changes in some growth parameters of Arabidopsis seedlings exposed to VOCs produced by a novel strain, L. macroides Xi9. In divided petri plate, only VOCs could influence plant growth. Six VOCs with relatively high peak areas were identified for strain Xi9. Linalool could reduce decay and increase antioxidant activities in blueberries (Wang et al. 2008). Heptadecane from actinomycete strain SPS-33 have the potential to inhibit mycelial growth and sporulation of C. fimbriata (Li et al. 2020). These VOCs could induce system resistance of plant, inhibit pathogens, and indirectly promote plants growth. However, the promotion effect of the identified compounds need further verification.
Studies have shown that VOCs released by bacteria can promote the production of auxin and/or other plant hormones that benefit plant development and growth (Timmusk et al. 1999; Velázquez-Becerra et al. 2011; Zhang et al. 2007). For bacterial VOCs with biological activity, without external auxin, which could facilitate plant growth via synthesis and transport auxin. Auxin plays an indispensible role in root growth and development including cell elongation, meristem size (Takatsuka and Umeda 2014; Qiu et al. 2020). In addition, lateral root growth and development also regulated by auxin (Bhalerao et al. 2002). The expression of genes encoding auxin response and transport were altered after exposure to VOCs of strain Xi9, such as the SAUR41 subfamily, ARF20, and PILS5. Consistently, the auxin content in the Xi9 group was significantly higher than control group. The SAURs comprised the largest family of early auxin-responsive genes, including 81 SAURs in Arabidopsis, which regulate a series of physiological, developmental, and cellular processes. SAUR41 and SAUR76 both positively mediate root growth (Ren and Gray 2015). Auxin efflux carrier family, PILS activity may influence the level of the endogenic auxin through intracellular metabolism and accumulation. The gene PILS5 negatively regulates the lateral root density (Barbez et al. 2012). The increase in auxin synthesis promoted advances in fresh weight, lateral roots and root hairs of Arabidopsis seedlings, but the primary root length did not increase (Fig. 2), necessitating further research.
As a macronutrient, nitrogen is necessary for plant growth and development normally (Hua et al. 2020). Nitrate (NO3−) is the most easily accessible form of nitrogen for plants (Zheng et al. 2013). NO3− reduction, absorption and assimilation are indispensable for plant growth (Williams and Miller 2001). Gene families NRT1 and NRT2 are critical for nitrate uptake and transport in Arabidopsis (Li et al. 2010). Mantelin et al. (2006) reported that a PGPB, Phyllobacterium brassicacearum STM196, mainly affects lateral root growth of Arabidopsis seedlings, which can change the plant nitrogen status. It was demonstrated that NRT2.5 and NRT2.6 benefit plant growth don’t depend on the N-sensing condition (Kechid et al. 2013). In our study, genes related to nitrogen assimilation and nitrate transport were up-regulated after exposure to VOCs of strain Xi9, such as NRT1.8, NRT2.1, NRT2.3, NRT2.5, NRT2.6 and NRT3.1. This result is similar to the growth-promoting mechanism of STM196. Whether the up-regulation of genes NRT2.5 and NRT2.6 are independent with the N-sensing condition in plant growth promotion require further verification. By detecting the NO3− and amino acid content of Arabidopsis seedlings, the Xi9 group showed higher levels than control group, further indicating that VOCs emitted by strain Xi9 can change the transcription level of NRT1 and NRT2 gene families and the nitrogen status of plants.
The cell wall is a continuously changing architecture that is engineered to regulate their biomechanical properties, promote growth and respond to various stimulus, such as plant hormones (Lewis et al. 2013; Sánchez-Rodríguez et al. 2010). VOCs of B. subtilius GB03 regulate cell wall-loosening proteins in Arabidopsis seedlings, wherein the genes encoding cell wall modification changed, showing an expression pattern that facilitates cell expansion. Several other genes encoding cell wall modification for reducing cell wall rigidity are up-regulated, leading to a degradation of cell wall (Zhang et al. 2007). Compared with the control group, the expression of genes encoding potential auxin-related expansion and pectate lyase were similarly up-regulated in the Xi9 group, such as At2g43890, EXPA12, EXPA16, and EXPA17: EXPA16 was expressed in the shoot, EXPA12 and EXPA17 were mainly expressed in the root (Lee and Kim 2013). Cell wall reshaping can regulate lateral root development and change cell elongation by altered genes transcription level associated with cell wall-modifying enzymes (Lewis et al. 2013). EXPA17 is thought to be significant for lateral roots (Lee and Kim 2013). As expected, after exposure to VOCs of strain Xi9 for 7 days, owing to cell wall reshaping, the cells of leaf elongated and lateral root numbers increased.
The root system is the main organ for plants to absorb nutrients and water from the soil. It is also the site of synthesis of a variety of metabolites, for instance, auxin and cytokinin. The plant root system represents significant flexibility in physiology and morphology to adapt to changes of environmental situations (Gutiérrez-Luna et al. 2010; Xun et al. 2020). In our result, VOCs emitted by L. macroides Xi9 not only promoted Arabidopsis seedlings growth but also modulated the root-system architecture. The number of lateral roots is closely related to the overall vitality of the roots and the utilization of nutrients and water, which are extremely important for plant growth and development. The root hair is the outward protrusion of the epidermal cells (ZhiMing et al. 2011), which readily to cling to the soil, and can increase the surface area of roots to absorb nutrients and effectively absorb nutrients from soil. The increase of lateral roots and root hairs may be related to the comprehensive results of several factors, or a single factor such as PILS, ANR1 and/or EXPA genes. The increase in lateral roots and root hairs can promote plant roots to absorb more nutrients from soil, promoting plant growth, including shoot and root.
Conclusion
We observed that VOCs from L. macroides Xi9 have plant growth promoting ability in Arabidopsis seedlings. Six VOCs from strain Xi9 were detected including three alkanes, two esters, and linalool. We explored differential expression of genes in Arabidopsis seedlings influenced by VOCs of L. macroides Xi9, mainly involved auxin synthesis and transport, cell wall modification and nitrogen absorption and transport. These findings indicated L. macroides Xi9 can facilitate plant growth by mediating nitrogen absorption, transport, auxin synthesis and cell wall modification. The results of this work provide a new insight in the interactions between VOCs of strain Xi9 and Arabidopsis seedlings in terms of nitrogen absorption, auxin synthesis and cell wall modification.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31770115, 32170133), the Tai-Shan Scholar Program from the Shandong Provincial Government.
Author contributions
YD and BD designed the study. DZ and JJ performed the laboratory work and analyzed the data. DZ wrote the manuscript. CW and KL revised the manuscript. YD, BD, and CW supported the study.
Funding
The National Natural Science Foundation of China (31770115) supported Prof. Yanqin Ding. The National Natural Science Foundation of China (32170133) supported Associate Prof. Chengqiang Wang.
Data availability
The transcriptome data used in the current study have been submitted to the NCBI Sequence Read Archive (SRA), and the accession number was PRJNA910295.
Declarations
Conflict of interest
All the authors declare no competing interests.
Ethical approval
The research does not involve human and animal subjects.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dongying Zhao, Email: zhaodongying4321@163.com.
Junhui Jiao, Email: jjh9506@163.com.
Binghai Du, Email: du_binghai@163.com.
Kai Liu, Email: liukai_1982@163.com.
Chengqiang Wang, Email: wangcq@sdau.edu.cn.
Yanqin Ding, Email: dyq@sdau.edu.cn.
References
- Anders S, Pyl PT, Huber W. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W (2012) Differential expression of RNA-Seq data at the gene level—the DESeq package, Heidelberg, Germany: European Molecular Biology Laboratory (EMBL) 10:f1000research
- Barbez E, Kubeš M, Rolčík J, Béziat C, Pěnčík A, Wang B, et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature. 2012;485:119–122. doi: 10.1038/nature11001. [DOI] [PubMed] [Google Scholar]
- Batista BD, Bonatelli ML, Quecine MC (2021) Fast screening of bacteria for plant growth promoting traits. In: The plant microbiome: Springer, pp 61–75. 10.1007/978-1-0716-1040-4_7 [DOI] [PubMed]
- Bhalerao RP, Eklöf J, Ljung K, Marchant A, Bennett M, Sandberg G. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 2002;29:325–332. doi: 10.1046/j.0960-7412.2001.01217.x. [DOI] [PubMed] [Google Scholar]
- Cao Y, Sun Q, Feng Z, Handique U, Wu J, Li M, Zhang R. First report of Pectobacterium parmentieri causing blackleg on potato in Inner Mongolia, China. Plant Dis. 2021 doi: 10.1094/PDIS-11-20-2502-PDN. [DOI] [PubMed] [Google Scholar]
- Chen W, Wang J, Huang D, Cheng W, Shao Z, Cai M, et al. Volatile organic compounds from Bacillus aryabhattai MCCC 1K02966 with multiple modes against Meloidogyne incognita. Molecules. 2021;27:103. doi: 10.3390/molecules27010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary DK, Kasotia A, Jain S, Vaishnav A, Kumari S, Sharma KP, et al. Bacterial-mediated tolerance and resistance to plants under abiotic and biotic stresses. J Plant Growth Regul. 2016;35:276–300. doi: 10.1007/s00344-015-9521-x. [DOI] [Google Scholar]
- Coorevits A, Dinsdale AE, Heyrman J, Schumann P, Van Landschoot A, Logan NA, et al. Lysinibacillus macroides sp. nov., nom. rev. Int J Syst Evol Micr. 2012;62:1121–1127. doi: 10.1099/ijs.0.027995-0. [DOI] [PubMed] [Google Scholar]
- Ferreira CM, Soares HM, Soares EV. Promising bacterial genera for agricultural practices: an insight on plant growth-promoting properties and microbial safety aspects. Sci Total Environ. 2019;682:779–799. doi: 10.1016/j.scitotenv.2019.04.225. [DOI] [PubMed] [Google Scholar]
- Gan Y, Bernreiter A, Filleur S, Abram B, Forde BG. Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 2012;53:1003–1016. doi: 10.1093/pcp/pcs050. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Luna FM, López-Bucio J, Altamirano-Hernández J, Valencia-Cantero E, De La Cruz HR, Macías-Rodríguez L. Plant growth-promoting rhizobacteria modulate root-system architecture in Arabidopsis thaliana through volatile organic compound emission. Symbiosis. 2010;51:75–83. doi: 10.1007/s13199-010-0066-2. [DOI] [Google Scholar]
- Hua Y-p, Zhou T, Huang J-y, Yue C-p, Song H-x, Guan C-y, et al. Genome-wide differential DNA methylation and miRNA expression profiling reveals epigenetic regulatory mechanisms underlying nitrogen-limitation-triggered adaptation and use efficiency enhancement in allotetraploid rapeseed. Int J Mol Sci. 2020;21:8453. doi: 10.3390/ijms21228453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar K, Bhattacharya R, Sharma A, Parmar K, Nath U, Nataraja KN, et al. Lysinibacillus macroides-mediated control of cellulose-producing morphotype of Salmonella. J Sci Food Agr. 2022;102:6491–6501. doi: 10.1002/jsfa.12016. [DOI] [PubMed] [Google Scholar]
- Kechid M, Desbrosses G, Rokhsi W, Varoquaux F, Djekoun A, Touraine B. The NRT 2.5 and NRT 2.6 genes are involved in growth promotion of Arabidopsis by the plant growth-promoting rhizobacterium (PGPR) strain Phyllobacterium brassicacearum STM 196. New Phytol. 2013;198:514–524. doi: 10.1111/nph.12158. [DOI] [PubMed] [Google Scholar]
- Kelen M, Demiralay EC, Şen S, Alsancak GÖ. Separation of abscisic acid, indole-3-acetic acid, gibberellic acid in 99 R (Vitis berlandieri x Vitis rupestris) and rose oil (Rosa damascena Mill.) by reversed phase liquid chromatography. Turk J Chem. 2004;28:603–610. [Google Scholar]
- Khan AL, Waqas M, Kang S-M, Al-Harrasi A, Hussain J, Al-Rawahi A, et al. Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J Microbiol. 2014;52:689–695. doi: 10.1007/s12275-014-4002-7. [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim J. EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 2013;54:1600–1611. doi: 10.1093/pcp/pct105. [DOI] [PubMed] [Google Scholar]
- Lemfack MC, Gohlke B-O, Toguem SMT, Preissner S, Piechulla B, Preissner R. mVOC 2.0: a database of microbial volatiles. Nucleic Acids Res. 2018;46:D1261–D1265. doi: 10.1093/nar/gkx1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Olex AL, Lundy SR, Turkett WH, Fetrow JS, Muday GK. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. Plant Cell. 2013;25:3329–3346. doi: 10.1105/tpc.113.114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-Y, Fu Y-L, Pike SM, Bao J, Tian W, Zhang Y, et al. The Arabidopsis nitrate transporter NRT1. 8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell. 2010;22:1633–1646. doi: 10.1105/tpc.110.075242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li B, Cai S, Zhang Y, Xu M, Zhang C, et al. Identification of rhizospheric actinomycete Streptomyces lavendulae sps-33 and the inhibitory effect of its volatile organic compounds against Ceratocystis fimbriata in postharvest sweet potato (Ipomoea batatas (L.) Lam.) Microorganisms. 2020;8:319. doi: 10.3390/microorganisms8030319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P-S, Kong W-L, Wu X-Q, Zhang Y. Volatile organic compounds of the plant growth-promoting rhizobacteria JZ-GX1 enhanced the tolerance of Robinia pseudoacacia to salt stress. Front Plant Sci. 2021 doi: 10.3389/fpls.2021.753332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-T, Lee C-C, Leu W-M, Wu J-J, Huang Y-C, Meng M. Fungicidal activity of volatile organic compounds emitted by Burkholderia gladioli strain BBB-01. Molecules. 2021;26:745. doi: 10.3390/molecules26030745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G-h, Liu B, Wang J-p, Che J-M, Chen Q-Q, Chen Z, et al. Genome sequence of type strain Lysinibacillus macroides DSM 54T. Genome Announc. 2015;3(6):e01271–e1315. doi: 10.1128/genomeA.01271-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed A, Muhammad Z, Ahmad H. Plant growth promoting bacteria: role in soil improvement, abiotic and biotic stress management of crops. Plant Cell Rep. 2018;37:1599–1609. doi: 10.1007/s00299-018-2341-2. [DOI] [PubMed] [Google Scholar]
- Mantelin S, Desbrosses G, Larcher M, Tranbarger TJ, Cleyet-Marel J-C, Touraine B. Nitrate-dependent control of root architecture and N nutrition are altered by a plant growth-promoting Phyllobacterium sp. Planta. 2006;223:591–603. doi: 10.1007/s00425-005-0106-y. [DOI] [PubMed] [Google Scholar]
- Mishra P, Mishra J, Arora NK. Plant growth promoting bacteria for combating salinity stress in plants: recent developments and prospects: a review. Microbiol Res. 2021;252:126861. doi: 10.1016/j.micres.2021.126861. [DOI] [PubMed] [Google Scholar]
- Park Y-S, Dutta S, Ann M, Raaijmakers JM, Park K. Promotion of plant growth by Pseudomonas fluorescens strain SS101 via novel volatile organic compounds. Biochem Bioph Res Co. 2015;461:361–365. doi: 10.1016/j.bbrc.2015.04.039. [DOI] [PubMed] [Google Scholar]
- Patten CL, Glick BR. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl Environ Microbiol. 2002;68:3795–3801. doi: 10.1128/AEM.68.8.3795-3801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pastrana J, Testillano PS, Barany I, Canto-Flick A, Álvarez-López D, Pijeira-Fernández G, et al. Endogenous auxin accumulation/localization during zygotic and somatic embryogenesis of Capsicum chinense Jacq. J Plant Physiol. 2021;258:153333. doi: 10.1016/j.jplph.2020.153333. [DOI] [PubMed] [Google Scholar]
- Qi G, Pan Z, Sugawa Y, Andriamanohiarisoamanana FJ, Yamashiro T, Iwasaki M, et al. Comparative fertilizer properties of digestates from mesophilic and thermophilic anaerobic digestion of dairy manure: focusing on plant growth promoting bacteria (PGPB) and environmental risk. J Mater Cycles Waste. 2018;20:1448–1457. doi: 10.1007/s10163-018-0708-7. [DOI] [Google Scholar]
- Qiu T, Qi M, Ding X, Zheng Y, Zhou T, Chen Y, et al. The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings. Ann Bot-Lond. 2020;125:805–819. doi: 10.1093/aob/mcz160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Gray WM. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant. 2015;8:1153–1164. doi: 10.1016/j.molp.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C-M, Farag MA, Hu C-H, Reddy MS, Kloepper JW, Paré PW. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004;134:1017–1026. doi: 10.1104/pp.103.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C-M, Farag MA, Hu C-H, Reddy MS, Wei H-X, Paré PW et al (2003) Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci 100 (8):4927–4932, P Natl Acad Sci USA 100: 4927–4932. 10.1073/pnas.0730845100 [DOI] [PMC free article] [PubMed]
- Sánchez-Rodríguez C, Rubio-Somoza I, Sibout R, Persson S. Phytohormones and the cell wall in Arabidopsis during seedling growth. Trends Plant Sci. 2010;15:291–301. doi: 10.1016/j.tplants.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Santoro MV, Zygadlo J, Giordano W, Banchio E. Volatile organic compounds from rhizobacteria increase biosynthesis of essential oils and growth parameters in peppermint (Mentha piperita) Plant Physiol Bioch. 2011;49:1177–1182. doi: 10.1016/j.plaphy.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Schöller CE, Gürtler H, Pedersen R, Molin S, Wilkins K. Volatile metabolites from actinomycetes. J Agr Food Chem. 2002;50:2615–2621. doi: 10.1021/jf0116754. [DOI] [PubMed] [Google Scholar]
- Sun S-W, Lin Y-C, Weng Y-M, Chen M-J. Efficiency improvements on ninhydrin method for amino acid quantification. J Food Compos Anal. 2006;19:112–117. doi: 10.1016/j.jfca.2005.04.006. [DOI] [Google Scholar]
- Syed-Ab-Rahman SF, Carvalhais LC, Chua ET, Chung FY, Moyle PM, Eltanahy EG, et al. Soil bacterial diffusible and volatile organic compounds inhibit Phytophthora capsici and promote plant growth. Sci Total Environ. 2019;692:267–280. doi: 10.1016/j.scitotenv.2019.07.061. [DOI] [PubMed] [Google Scholar]
- Tahir HA, Gu Q, Wu H, Raza W, Hanif A, Wu L, et al. Bacillus volatiles adversely affect the physiology and ultra-structure of Ralstonia solanacearum and induce systemic resistance in tobacco against bacterial wilt. Sci Rep-UK. 2017;7:1–15. doi: 10.1038/srep40481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir HAS, Gu Q, Wu H, Niu Y, Huo R, Gao X. Plant growth promotion by volatile organic compounds produced by Bacillus subtilis SYST2. Front Microbiol. 2017;8:171. doi: 10.3389/fmicb.2017.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka H, Umeda M. Hormonal control of cell division and elongation along differentiation trajectories in roots. J Exp Bot. 2014;65:2633–2643. doi: 10.1093/jxb/ert485. [DOI] [PubMed] [Google Scholar]
- Tilocca B, Cao A, Migheli Q. Scent of a killer: microbial volatilome and its role in the biological control of plant pathogens. Front Microbiol. 2020;11:41. doi: 10.3389/fmicb.2020.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk S, Nicander B, Granhall U, Tillberg E. Cytokinin production by Paenibacillus polymyxa. Soil Biol Biochem. 1999;31:1847–1852. doi: 10.1007/s12088-009-0008-y. [DOI] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez-Becerra C, Macías-Rodríguez LI, López-Bucio J, Altamirano-Hernández J, Flores-Cortez I, Valencia-Cantero E. A volatile organic compound analysis from Arthrobacter agilis identifies dimethylhexadecylamine, an amino-containing lipid modulating bacterial growth and Medicago sativa morphogenesis in vitro. Plant Soil. 2011;339:329–340. doi: 10.1007/s11104-010-0583-z. [DOI] [Google Scholar]
- Wang CY, Wang SY, Chen C. Increasing antioxidant activity and reducing decay of blueberries by essential oils. J Agr Food Chem. 2008;56(10):3587–3592. doi: 10.1021/jf7037696. [DOI] [PubMed] [Google Scholar]
- Wang C, Zhao D, Qi G, Mao Z, Hu X, Du B, et al. Effects of Bacillus velezensis FKM10 for promoting the growth of Malus hupehensis Rehd. and inhibiting Fusarium verticillioides. Front Microbiol. 2020;10:2889. doi: 10.3389/fmicb.2019.02889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisskopf L, Schulz S, Garbeva P. Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat Rev Microbiol. 2021;19:391–404. doi: 10.1038/s41579-020-00508-1. [DOI] [PubMed] [Google Scholar]
- Williams L, Miller A. Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu Rev Plant Biol. 2001;52:659–688. doi: 10.1146/annurev.arplant.52.1.659. [DOI] [PubMed] [Google Scholar]
- Xu N, Wang R, Zhao L, Zhang C, Li Z, Lei Z, et al. The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell. 2016;28:485–504. doi: 10.1105/tpc.15.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun Q, Wu Y, Li H, Chang J, Ou Y, He K, et al. Two receptor-like protein kinases, MUSTACHES and MUSTACHES-LIKE, regulate lateral root development in Arabidopsis thaliana. New Phytol. 2020;227:1157–1173. doi: 10.1111/nph.16599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kim M-S, Krishnamachari V, Payton P, Sun Y, Grimson M, et al. Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta. 2007;226:839. doi: 10.1007/s00425-007-0530-2. [DOI] [PubMed] [Google Scholar]
- Zheng D, Han X, An Y, Guo H, Xia X, Yin W. The nitrate transporter NRT2. 1 functions in the ethylene response to nitrate deficiency in Arabidopsis. Plant Cell Environ. 2013;36:1328–1337. doi: 10.1111/pce.12062. [DOI] [PubMed] [Google Scholar]
- ZhiMing Y, Bo K, XiaoWei H, ShaoLei L, YouHuang B, WoNa D, et al. Root hair-specific expansins modulate root hair elongation in rice. Plant J. 2011;66:725–734. doi: 10.1111/j.1365-313x.2011.04533.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome data used in the current study have been submitted to the NCBI Sequence Read Archive (SRA), and the accession number was PRJNA910295.