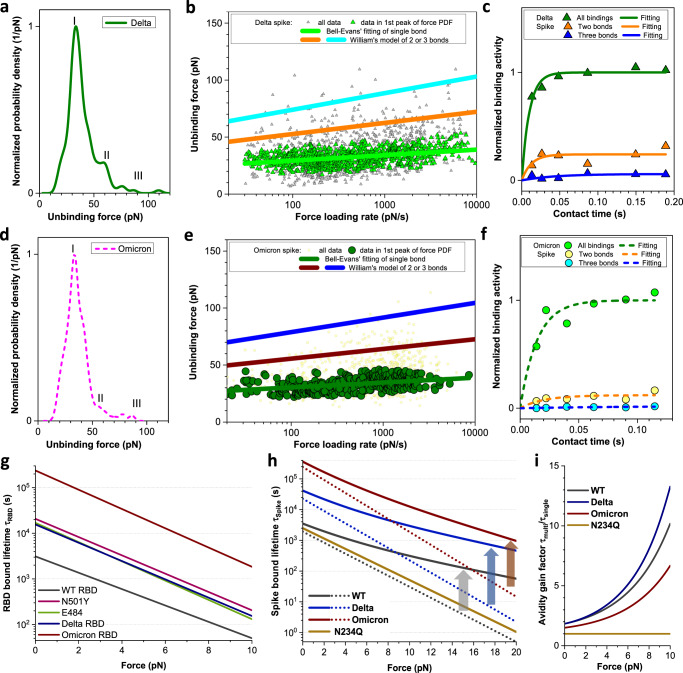
Fig. 5. Mutations and avidity amplification lead to the prolongation of Spike binding to ACE2.
a, d Experimental PDFs from measured dissociation forces between Spike and ACE2. Force maxima for the Spike trimer Delta (a) and Omicron (B.1.1.529) (d) variants correspond to the simultaneous dissociation of one (I), two (II), or three (III) bonds. b, e Dependence of dissociation forces on the force loading rate. Data of the first peak (b, green triangles for Delta variant and e olive circles for Omicron (B.1.1.529)) were fitted with the Bell–Evans theory (b, green line for Delta variant and e, olive line for Omicron (B.1.1.529)). The Markov binding model for two and three bonds are computed and shown as orange and cyan lines for the Delta Spike (b) and wine and blue lines for Omicron (B.1.1.529) Spike (e), respectively. c, f Dependence of binding activity on contact time between AFM tip and cellular surfaces for the Delta Spike (c olive triangle) and the Omicron (B.1.1.529) Spike (f green circles). Ratio of force curves containing two (c, orange triangles for Delta and f, yellow circles for Omicron (B.1.1.529) Spikes) and three (c blue triangles for Delta and f cyan circles for Omicron (B.1.1.529) Spikes) bond breakages are shown and fitted with pseudo-first-order kinetics (c for Delta and f for Omicron (B.1.1.529) Spike; orange lines for two bonds and blue lines for three bonds). g Lifetime of the RBD:ACE2 complex under force for the Wuhan reference strain (WT), the indicated mutants, and variants. h Lifetime of the Spike:ACE2 complex at increasing external force for WT and the indicated variants. Dotted or solid lines represent one RBD or full trimeric Spike. Arrows (gray for WT, blue for Delta, brown for Omicron) indicate lifetime increase arising from multivalent Spike binding. i Ratio of bound lifetimes for RBD triple bonds compared to individual RBD bonds, termed as avidity gain factor τmulti/τ1Spike. The lifetimes of WT and the indicated variant Spikes were calculated according to the kinetic model of Williams except for N234Q, where only a single RBD binds. Source data are provided as a Source Data file.