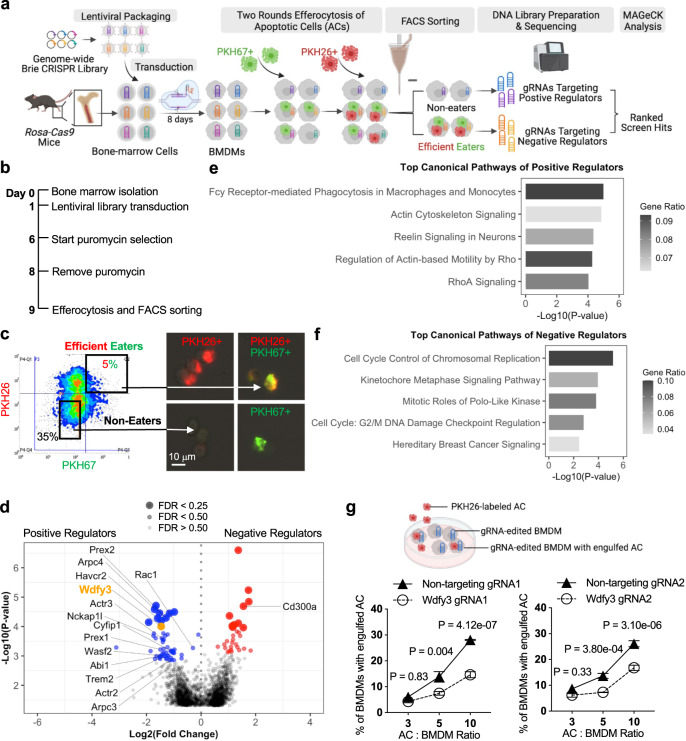
Fig. 1. A pooled, fluorescence-activated cell sorting (FACS)-based genome-wide CRISPR knockout screen in primary mouse macrophages identified known and novel regulators of macrophage efferocytosis.
a Schematics of the CRISPR screen workflow. b Timeline of bone marrow isolation, lentiviral library transduction, puromycin selection, efferocytosis, and cell sorting. c Visualization of gating strategy for separation of non-eaters and efficient eaters. Successful separation was confirmed by fluorescent microscopy. d Volcano plot highlights the top-ranked screen hits that are known positive and negative regulators of macrophage efferocytosis. e Canonical pathways enriched in top-ranked positive regulators by Ingenuity Pathway Analysis (IPA). f Canonical pathways enriched in top-ranked negative regulators by IPA. g Validation of Wdfy3 as a positive regulator required for macrophage efferocytosis (n = 4 independent experiments). Data are presented as mean ± SEM. Two-sided P values were determined by a two-way ANOVA with Tukey’s multiple comparisons test in panel g. ACs, apoptotic cells; BMDMs, bone-marrow-derived macrophages.