FIGURE 4.
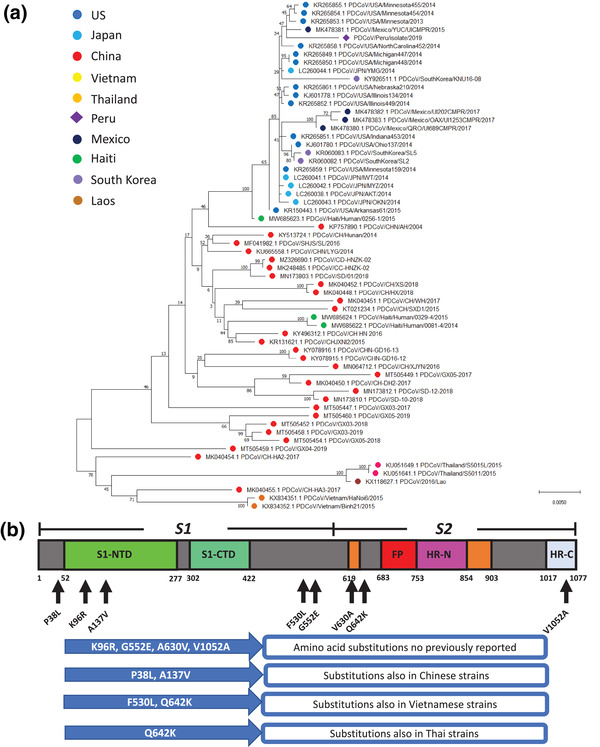
Phylogenetic analysis of S gene reveals that Peruvian PDCoV strain has a close relationship with a Mexican strain within the North American phylogroup. The evolutionary history was inferred by using the maximum likelihood method and general time reversible model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. A discrete gamma distribution was used to model evolutionary rate differences among. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 62 nucleotide sequences. Evolutionary analyses were conducted in MEGA X (a). Schematic representation of the porcine deltacoronavirus S protein highlighting the amino acid substitutions found in the present study. Black arrows represent the site of these substitutions. A multiple amino acid sequence alignment of S protein was performed using Clustal W in MEGA X. Eight amino acid modifications were detected within the Peruvian sequence compared to others (b)
