Abstract
Human DCs have been divided into several subsets based on their phenotype and ontogeny. Recent high throughput single‐cell methods have revealed additional heterogeneity within human DC subsets, and new subpopulations have been proposed. In this review, we provide an updated view of the human DC subsets and of their ontogeny supported by recent clinical studies . We also summarize their main characteristics including their functional specialization.
Keywords: Human, dendritic cell, subsets
We provide an updated view of the human dendritic cell subsets and of their ontogeny. We also summarize their main characteristics including their functional specialization.
Introduction
The heterogeneity of human DC has been described initially based on their surface phenotype [1] and transcriptome [2]. Recently, high‐dimensional methods have refined the description of human DC subsets by identifying previously overlooked DC populations. The caveats and challenges of using single‐cell transcriptomics for defining novel DC subsets, in particular distinguishing bona fide subsets from transitory cellular states, have already been discussed elsewhere [3]. The consensus nomenclature for DC is based on ontogeny [4], that is, a DC population is considered to represent a distinct subset if it possesses a specific developmental pathway including distinct transcription factors enforcing their lineage commitment and/or identity. Following this nomenclature, human DC can be classified into classical DC (cDC) type 1 (cDC1), cDC type 2 (cDC2), DC3, plasmacytoid DC (pDC), and monocyte‐derived DC (mo‐DC). As discussed in this review, there is sufficient evidence that these DC populations develop along distinct pathways. A population of AXL+ SIGLEC6+ DC has also been described, but whether it represents a DC subset remains unclear (discussed in Section “Characteristics of human transitional AXL+ SIGLEC6+ DC”). Langerhans cells, which are classified as a population of skin macrophages, will not be discussed in this review.
Human DC subset identity is imprinted by their ontogeny, as DC subsets from distinct organs display a common transcriptomic program [5] and shared phenotypic markers (Table 1). In addition, there is a level of tissue imprinting, as specific signatures exist in mucosal tissue DC, for instance, the expression of CD103 for cDC1 or CD1a for cDC2 [5, 6, 7, 8]. Of note, there is also significant interindividual variation in the phenotype of cDC2 [7]. Recent single‐cell RNA‐seq (scRNA‐seq) studies have also shown that all human DC subsets express a common activation program upon maturation, both homeostatic and induced by inflammatory stimuli [3].
Table 1.
Phenotypic markers of human DC subsets
| Markers | cDC1 | cDC2 | pDC | DC3 | Mo‐DC | CD14+ monocyte |
|---|---|---|---|---|---|---|
| CADM1 | + | − | − | − | − | − |
| CD11c | + | ++ | − | ++ | ++ | ++ |
| CD123 | − | − | + | − | − | − |
| CD14 | − | − | − | Low to + | + | + |
| CD141 | ++ | + | − | + | + | − |
| CD163 | − | − | − | + | − | − |
| CD172a | − | + | − | ? | + | + |
| CD1a | − | Tissue‐dependent | − | − | + | − |
| CD1c | − | + | − | + | + | − |
| CD226 | + | − | − | − | + | − |
| CD303 | − | − | + | − | − | − |
| CD304 | − | − | + | − | − | − |
| CD5 | − | Low to + | − | − | ? | − |
| CD64 | − | Tissue‐dependent | − | ? | + | + |
| CD88 | − | − | − | − | + | ++ |
| Clec10A | − | + | − | + | + | − |
| Clec9A | + | − | − | − | − | − |
| FcεRI | − | + | − | + | + | − |
| S100A8/A9 | − | − | − | + | + | + |
| XCR1 | + | − | − | − | − | − |
Ontogeny of human DC subsets
In vitro differentiation models have provided insights into the ontogeny of human DC subsets (Figure 1). pDC and pre‐cDC arise from a common dendritic cell progenitor (CDP) downstream of a IRF8high GMDP (granulocyte‐monocyte‐DC progenitor) [9, 10, 11]. A series of studies have shown that pDC in the mouse possess a dual origin, with pre‐pDC deriving from CDP or from a lymphoid progenitor [12]. Whether the same holds true for human remains to be determined. Of note, human multipotent lymphoid progenitors have also been shown to give rise to cDC, with a bias toward cDC1 [13]. Pre‐cDC display heterogeneity at the transcriptomic level as shown by single‐cell RNA‐seq, and are precommitted to become cDC1 or cDC2 [14, 15]. IRF8low GMDP gives rise to monocytes and DC3 along separate routes [11]. Finally, monocytes differentiate into macrophages or mo‐DC via two distinct developmental pathways [16].
Figure 1.
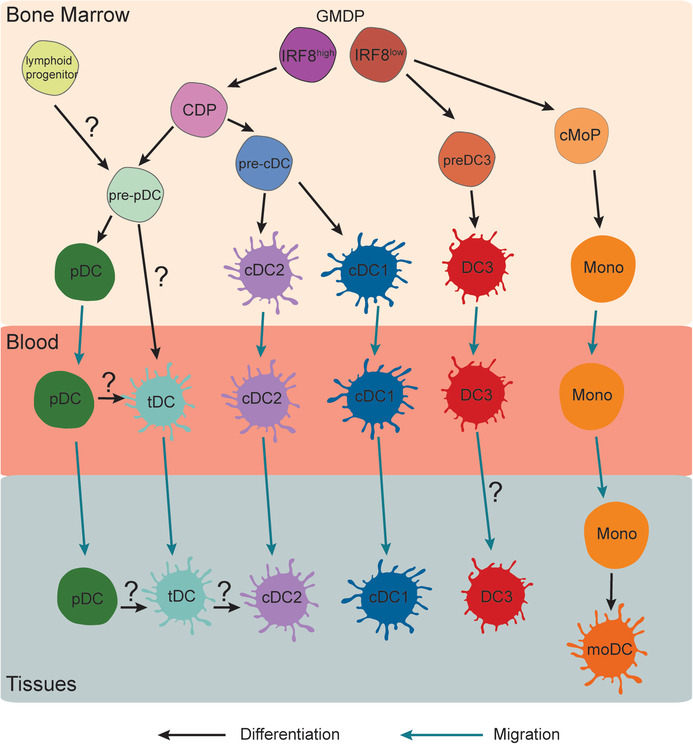
Ontogeny of human DC subsets. DC precursors originate from the bone marrow. pDC, cDC1, cDC2, and DC3 develop from precursors distinct from the other DC lineages. moDC are differentiated from monocytes in peripheral tissues. The developmental pathway of tDC remains to be better characterized. Cell differentiation is indicated in black and cell migration in blue. Questions marks indicate aspects that remain unclear. CDP, common DC progenitor; cMoP, common monocyte precursor; GMDP, granulocyte‐monocyte and DC progenitor, mono, monocyte; moDC, monocyte‐derived DC; tDC, transitional AXL+ SIGLEC6+ DC.
Studies of primary immunodeficiencies have confirmed the essential role of IRF8 for human DC development in vivo. Patients bearing a dominant negative IRF8 mutation (resulting in reduced activity) have severely reduced numbers of pDC, cDC1, and cDC2 [17, 18, 19, 20], but maintain DC3 numbers [11]. By contrast, patients with a total loss‐of‐function IRF8 mutation lack monocyte and DC development entirely [11].
Other transcription factors involved in pDC development include SpiB and E2‐2/Tcf4 as shown in in vitro models [21, 22]. Patients with a deficiency in IKZF1 (encoding Ikaros) have reduced circulating pDC, showing a role for Ikaros in pDC development in vivo [23]. By contrast, patients with a loss‐of‐function mutation or deletion of E2‐2 have normal numbers of pDC but their phenotype and function are altered, suggesting a role for E2‐2/Tcf4 in a late stage of pDC differentiation in vivo [24].
Activation of the Notch pathway inhibits pDC development but is critical for cDC1 differentiation in in vitro culture systems [25, 26, 27].
Finally, mo‐DC differentiation is dependent on IRF4, Blimp‐1, aryl hydrocarbon receptor and NCOR2, as evidenced in in vitro models [16, 28].
Characteristics of human cDC1
cDC1 are found in peripheral tissues and in lymphoid organs. Analysis of mucosal tissues and associated draining LNs has suggested that mucosal cDC1 have lower migratory ability than cDC2 [6]. In lymphoid organs, cDC1 are dispersed in the T‐cell areas [6, 29, 30].
Blood, lymphoid organ, and lung cDC1 have been shown ex vivo to stimulate naïve CD4 T‐cell polarization into both Th1 and Th2 cells [29, 30, 31, 32].
Blood, skin, and lymphoid organ cDC1 are efficient cross‐presenting cells, with a superior ability for the cross‐presentation of necrotic cell‐associated material [8, 30, 31, 33, 34, 35, 36, 37]. In ex vivo assays, they also promote the differentiation of cytotoxic CD8 T cells [38, 39].
In terms of cytokine secretion, cDC1 are specialized for the production of type III IFN [38, 40, 41].
Characteristics of human cDC2
cDC2 are found in peripheral tissues and in lymphoid organs, where they are enriched at the border of T–B‐cell zones [6, 29]. A recent scRNA‐seq study has reported two populations of cDC2 (Clec10A+ and Clec10A−) present in the human spleen, but not in the blood [42]. The significance of this finding is unclear, as these populations have not yet been observed by others. Whether Clec10A+ and Clec10A− cDC2 constitute distinct subsets or different cellular states remains unclear. In addition, their phenotype may be imprinted by a particular tissue microenvironment and their presence in other organs and tissues has to be confirmed.
Similar to cDC1, blood, lymphoid organ, skin, and lung cDC2 can induce ex‐vivo polarization of naïve CD4 T cells into Th1 and Th2 cells [29, 30, 31, 32, 43]. Blood and lung cDC2 have a superior ability for the induction of Th17 responses, which is likely due to their ability to secrete IL‐23 [44, 45]. Blood, lymphoid organ, lung, and skin cDC2 are also the most potent inducers of T‐follicular helper (Tfh) cells [29, 46, 47], due to their higher expression of Activin A and OX40‐ligand [29, 47]. Consistent with their role in Tfh polarization, lung cDC2 are recruited to tertiary lymphoid organs [47].
In the blood and lymphoid organs, cDC2 are as efficient as cDC1 for the cross‐presentation of soluble protein antigens ex vivo [30, 33, 34, 37]. In addition, they can stimulate the differentiation of cytotoxic CD8 T cells [38, 39].
Regarding cytokine secretion, in addition to IL‐23 and activin A, cDC2 are the most efficient for the production of IL‐12p70 [29, 33, 38].
Finally, intestinal cDC2 are superior to cDC1 for the induction of CD4 Treg, due to their higher expression of integrin αvβ8 which is essential for generating bioavailable TGF‐β [48].
Characteristics of human pDC
In the steady state, pDC are present in lymphoid organs but not in peripheral tissues.
The most characteristic feature of pDC is their specialization for the production of type I IFN upon activation [49].
Blood and lymphoid organ pDC are less efficient than cDC for the stimulation of naïve CD4 T cells in the steady state [29]. However, they can become potent stimulators of CD4 T cells after ex‐vivo activation, and are able to induce Th1 polarization [49]. It was reported that, upon ex‐vivo activation, only a subpopulation of pDC would become APCs, while others would be specialized for the secretion of type I IFN [50]. The physiological relevance of this observation remains unclear. Of note, pDC recognize virus‐infected cells via cell‐cell contact [51, 52, 53], which triggers long‐lasting IFN responses without the emergence of an antigen‐presenting population [51].
In the blood and lymphoid organs, pDC possess the ability to cross‐present soluble, cell‐associated or viral antigens in ex‐vivo assays [34, 54, 55, 56, 57, 58]. However, they are poor stimulators of cytotoxic CD8 T‐cell differentiation [39].
pDC are also able to induce CD4 Treg via their high expression of ICOS‐ligand [59] or of IDO, an enzyme that catabolizes tryptophan degradation [60].
Characteristics of human DC3
DC3 were initially identified in the blood by scRNA‐seq analysis [61]. They express a mixed cDC2‐monocyte transcriptomic and phenotypic profile (Table 1). They are best characterized by their coexpression of CD1c and CD163 [11, 62]. They have also been evidenced in the BM [11] and a population with characteristics of DC3 has been observed in oropharyngeal carcinomas [63] and in psoriatic skin [64], but the presence of DC3 in lymphoid organs and peripheral tissues remains to be better characterized. Of note, the term “DC3” has been used to refer to a DC population identified in multiple tumor samples, which should not be mistaken for DC3, as they actually correspond to mature DC [65].
Circulating DC3 are increased in the blood of systemic lupus erythematosus patients and of melanoma patients [62, 66]. DC3 are also specifically increased in the blood of severe COVID‐19 patients [67, 68]. Their potential role in the physiopathology of these diseases is unclear.
Blood DC3 are efficient for the stimulation of naïve CD4 T cells ex vivo [61, 62, 69]. DC3 have been reported to preferentially induce Th17 [62] or Th1 [69] polarization, depending on the study.
DC3 can also stimulate the proliferation of naïve CD8 T cells and their expression of maturation markers [69], but whether they can actually cross‐present antigens remains to be determined. It has also been proposed that DC3 have a superior ability to induce tissue‐resident memory T cells, as DC3 stimulate the expression on CD8 T cells of tissue‐homing molecule CD103 [69], and highly express upon type I IFN exposure the costimulatory molecule GITRL [70], which is important for the formation of tissue‐resident memory T cells.
Regarding cytokine production, DC3 are able to secrete IL‐12p70 and IL‐23, similar to cDC2, as well as large amounts of IL‐1β, similar to monocytes [11, 69].
Characteristics of human mo‐DC
mo‐DC share numerous phenotypic markers with monocyte‐derived macrophages, and it can be difficult to distinguish the two cell types from one another [71]. A key feature of mo‐DC is their dendritic morphology, similar to that of classical DC [72, 73, 74], whereas macrophages show a large cytoplasm containing numerous phagocytic vacuoles. Another typical characteristic of mo‐DC compared to macrophages is their superior ability to stimulate naïve T‐cell activation [39, 73], but this is not always possible to assess due to the challenges associated with cell isolation from human clinical samples. mo‐DC also express DC‐related transcriptomic signatures [39, 74, 75, 76], including transcription factors related to their molecular ontogeny such as IRF4 [16].
mo‐DC have been described in clinical samples both in steady state and inflammatory context. mo‐DC are present in steady‐state peritoneum [72], nondiseased intestine [75, 76], and lungs [77]. A population of CD14+ DC in the steady‐state skin is also believed to be monocyte‐derived [78, 79]. “Inflammatory” mo‐DC are also found in skin from atopic dermatitis and psoriasis patients [80, 81], pleural effusions from tuberculosis patients [82], peritoneal ascites from cancer patients [73], synovial fluid from rheumatoid arthritis patients [73], and intestinal lamina propria of Crohn's disease patients [83]. Finally, cells with phenotypic features of mo‐DC have been observed in breast [74], colorectal [84], lung [84, 85, 86] cancers, and melanoma‐draining LNs [87].
mo‐DC from clinical samples efficiently stimulate naïve CD4 T‐cell proliferation ex vivo and preferentially induce Th17 cells [73, 82] or Th1 cells [76, 81] depending on the context. mo‐DC from skin, synovial fluid, and peritoneal ascites have also been reported to efficiently induce Tfh polarization [29, 30, 43].
Peritoneal mo‐DC can cross‐present soluble and particulate antigens [39, 72], but use a nonconventional intracellular pathway dependent on lysosomal proteases [39]. mo‐DC are also efficient for inducing the differentiation of effector cytotoxic CD8 T cells [39].
Similar to cDC2, mo‐DC have been shown to be specialized for the secretion of IL‐23 [73, 82] and IL‐12p70 [39].
Characteristics of human transitional AXL+ SIGLEC6+ DC
AXL+ SIGLEC6+ DC were identified in the blood by scRNA‐seq analysis as a subpopulation of CD123+ DC [15, 61]. They have also been evidenced in lymphoid organs but not in steady‐state peripheral tissues [7], and are recruited to inflamed skin and lungs [88, 89].
AXL+ SIGLEC6+ DC display a mixed pDC‐cDC transcriptomic and phenotypic profile, however, they are closer to cDC2 functionally. They are efficient for stimulating CD4 T cells ex vivo, and do not secrete type I IFN [7, 15, 51, 61, 90, 91].
Whether AXL+ SIGLEC6+ DC represent a bona fide DC subset or an intermediate population remains to be confirmed (Figure 1). Because they can differentiate into cDC2 in culture systems [15, 61, 92], they have been proposed to be DC precursors or a “transitional” population between pDC and cDC2. Their molecular ontogeny remains unclear and might be shared with pDC. AXL+ SIGLEC6+ DC are decreased in the blood of patients with a mutation in TCF4 [15], suggesting a role for E2‐2/TCF4 in their differentiation. AXL+ SIGLEC6+ DC also highly express the transcription factors BCL11A, RUNX2, and SPIB, which are involved in pDC development [91].
Functional specialization of DC subsets
Numerous studies using genetic mouse models have identified specific functions for murine DC subsets and contributed to the concept of functional specializations of DC subsets and their “division of labor” [12, 93, 94]. While DC subsets are conserved between mice and humans [4], their functional properties are not always similar (Table 2). In particular, the ability to cross‐present antigens seems to be less restricted in human DC compared to murine DC subsets. However, DC subsets display distinct abilities for antigen uptake, with pDC being inefficient for engulfing large particles and cDC1 being superior for capturing necrotic cells, leading to some specialization in the type of antigen being actually cross‐presented in vivo. Another important aspect is their in‐situ localization. While different subsets may display similar abilities ex vivo, they may actually play complementary roles in an in‐vivo immune response due to their distinct localization or migratory capacity. For instance, monocytes are massively recruited during inflammation, and mo‐DC will outnumber other DC populations in the inflamed tissue. With their large array of functional properties (Table 2), mo‐DC could efficiently restimulate effector T cells which have been primed by cDC in lymphoid organs, or resident memory T cells being reactivated directly in the tissue.
Table 2.
Functional properties of mouse and human DC subsets
| Function | cDC1 | cDC2 | pDC | DC3 | Mo‐DC | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cross‐presentation | Yes | Yes | No | Yes | No | Yes | ? | Yes | Yes |
| Presentation on MHC II | Yes | Yes | Yes | Yes | Limited | Yes | Yes | Yes | Yes |
| Induction of cytotoxic CD8 T cells | Yes | Yes | No | Yes | No | Limited | ? | Yes | Yes |
| Induction of Th1 cells | Yes | Yes | No | Yes | No | Yes | Yes | Yes | Yes |
| Induction of Th2 cells | No | Yes | Yes | Yes | No | No | No | ? | ? |
| Induction of Th17 cells | No | No | Yes | Yes | No | No | Yes | Yes | Yes |
| Induction of Tfh cells | No | No | Yes | Yes | No | No | ? | ? | Yes |
| Induction of Treg cells | Yes | No | No | Yes | Yes | Yes | ? | ? | ? |
| Secretion of IL12p70 | Yes | Limited | No | Yes | No | No | Yes | Yes | Yes |
| Secretion of IL23 | No | No | Yes | Yes | No | No | Yes | Yes | Yes |
| Secretion of type I interferon | No | No | No | No | Yes | Yes | No | No | No |
| Secretion of type III interferon | Yes | Yes | No | No | Yes | Yes | ? | No | No |
The main immune functions of DC subsets are summarized, for humans and mice DC for comparison. Mouse DC characteristics are in blue, human DC in orange. For mouse DC functions, see details in recent reviews [12, 93, 94]. Question marks indicate that this function has not been reported yet in the literature. Tfh, T follicular helper, Treg, T regulatory.
In addition, when murine and human DC counterparts exert the same function, the precise molecular mechanisms involved can be different. This is the case for Tfh induction by cDC2, which relies on the production by DC of IL12p70 and Activin A in humans, but not in mice [29, 95]. Caution should, therefore, be exercised when extrapolating results from mice models to the human situation, and functional properties, including key molecular aspects, should be confirmed using human DC isolated from relevant tissues.
Conclusions and perspectives
Recent scRNA‐seq studies have unraveled underappreciated heterogeneity within historical DC subsets. Based on their distinct ontogeny, human DC can now be divided into cDC1, cDC2, DC3, pDC, and mo‐DC. Determining whether Clec10A+ and Clec10A− cDC2 and transitional AXL+ SIGLEC6+ DC represent additional DC subsets, cell states, or progenitors requires further investigation. A better characterization of the functional properties of DC3 and AXL+ SIGLEC6+ DC is also needed to understand their potential specialization compared to cDC2. Refined sets of markers should be used in future studies to distinguish and accurately identify human DC subsets, which will be essential for addressing their respective contributions to health and diseases.
Conflict of interest
The author declares no commercial or financial conflict of interest.
Author Contribution
ES wrote the manuscript.
Abbreviations
- cDC
classical DC
- CDP
common dendritic cell progenitor
- mo‐DC
monocyte‐derived DC
- pDC
plasmacytoid DC
- scRNA‐seq
single‐cell RNA‐seq
- Tfh
T‐follicular helper
Acknowledgment
This work was funded by INSERM.
Correction added on 28 June 2022, after first online publication: The copyright line was changed.
References
- 1. Dzionek, A. , Fuchs, A. , Schmidt, P. , Cremer, S. , Zysk, M. , Miltenyi, S. , Buck, D. W. et al., BDCA‐2, BDCA‐3, and BDCA‐4: three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. 2000. 165: 6037–6046. [DOI] [PubMed] [Google Scholar]
- 2. Robbins, S. H. , Walzer, T. , Dembélé, D. , Thibault, C. , Defays, A. , Bessou, G. , Xu, H. et al., Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome‐wide expression profiling. Genome Biol. 2008. 9: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Villar, J. and Segura, E. , Decoding the heterogeneity of human dendritic cell subsets. Trends Immunol. 2020. 41: 1062–1071. [DOI] [PubMed] [Google Scholar]
- 4. Guilliams, M. , Ginhoux, F. , Jakubzick, C. , Naik, S. H. , Onai, N. , Schraml, B. U. , Segura, E. et al., Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 2014. 14: 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heidkamp, G. F. , Sander, J. , Lehmann, C. H. K. , Heger, L. , Eissing, N. , Baranska, A. , Lühr, J. J. et al., Human lymphoid organ dendritic cell identity is predominantly dictated by ontogeny, not tissue microenvironment. Sci Immunol. 2016. 1. [DOI] [PubMed] [Google Scholar]
- 6. Granot, T. , Senda, T. , Carpenter, D. J. , Matsuoka, N. , Weiner, J. , Gordon, C. L. , Miron, M. et al., Dendritic Cells Display Subset and Tissue‐Specific Maturation Dynamics over Human Life. Immunity. 2017. 46: 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alcántara‐Hernández, M. , Leylek, R. , Wagar, L. E. , Engleman, E. G. , Keler, T. , Marinkovich, M. P. , Davis, M. M. et al., High‐Dimensional Phenotypic Mapping of Human Dendritic Cells Reveals Interindividual Variation and Tissue Specialization. Immunity. 2017. 47: 1037–1050.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haniffa, M. , Shin, A. , Bigley, V. , McGovern, N. , Teo, P. , See, P. , Wasan, P. S. et al., Human tissues contain CD141hi cross‐presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012. 37: 60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee, J. , Breton, G. , Oliveira, T. Y. K. , Zhou, Y. J. , Aljoufi, A. , Puhr, S. , Cameron, M. J. et al., Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J. Exp. Med. 2015. 212: 385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Breton, G. , Lee, J. , Zhou, Y. J. , Schreiber, J. J. , Keler, T. , Puhr, S. , Anandasabapathy, N. et al., Circulating precursors of human CD1c+ and CD141+ dendritic cells. J. Exp. Med. 2015. 212: 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cytlak, U. , Resteu, A. , Pagan, S. , Green, K. , Milne, P. , Maisuria, S. , McDonald, D. et al., Differential IRF8 transcription factor requirement defines two pathways of dendritic cell development in humans. Immunity. 2020. 53: 353–370.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderson, D. A. , Dutertre, C. ‐A. , Ginhoux, F. and Murphy, K. M. , Genetic models of human and mouse dendritic cell development and function. Nat. Rev. Immunol. 2021. 21: 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Helft, J. , Anjos‐Afonso, F. , van der Veen, A. G. , Chakravarty, P. and Bonnet, D. , Reis e Sousa C. Dendritic cell lineage potential in human early hematopoietic progenitors. Cell Rep. 2017. 20: 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Breton, G. , Zheng, S. , Valieris, R. , Tojal da Silva, I. , Satija, R. and Nussenzweig, M. C. , Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. J. Exp. Med. 2016. 213: 2861–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. See, P. , Dutertre, C. ‐A. , Chen, J. , Günther, P. , McGovern, N. , Irac, S. E. , Gunawan, M. et al., Mapping the human DC lineage through the integration of high‐dimensional techniques. Science. 2017. 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goudot, C. , Coillard, A. , Villani, A. ‐C. , Gueguen, P. , Cros, A. , Sarkizova, S. , Tang‐Huau, T. ‐L. et al., Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages. Immunity. 2017. 47: 582–596.e6. [DOI] [PubMed] [Google Scholar]
- 17. Bigley, V. , Maisuria, S. , Cytlak, U. , Jardine, L. , Care, M. A. , Green, K. , Gunawan, M. et al., Biallelic interferon regulatory factor 8 mutation: A complex immunodeficiency syndrome with dendritic cell deficiency, monocytopenia, and immune dysregulation. J. Allergy Clin. Immunol. 2018. 141: 2234–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hambleton, S. , Salem, S. , Bustamante, J. , Bigley, V. , Boisson‐Dupuis, S. , Azevedo, J. , Fortin, A. et al., IRF8 mutations and human dendritic‐cell immunodeficiency. N. Engl. J. Med. 2011. 365: 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bigley, V. , Haniffa, M. , Doulatov, S. , Wang, X. ‐N. , Dickinson, R. , McGovern, N. , Jardine, L. et al., The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J. Exp. Med. 2011. 208: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kong, X. ‐F. , Martinez‐Barricarte, R. , Kennedy, J. , Mele, F. , Lazarov, T. , Deenick, E. K. , Ma, C. S. et al., Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat. Immunol. 2018. 19: 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagasawa, M. , Schmidlin, H. , Hazekamp, M. G. , Schotte, R. and Blom, B. , Development of human plasmacytoid dendritic cells depends on the combined action of the basic helix‐loop‐helix factor E2‐2 and the Ets factor Spi‐B. Eur. J. Immunol. 2008. 38: 2389–2400. [DOI] [PubMed] [Google Scholar]
- 22. Schotte, R. , Nagasawa, M. , Weijer, K. , Spits, H. and Blom, B. , The ETS transcription factor Spi‐B is required for human plasmacytoid dendritic cell development. J. Exp. Med. 2004. 200: 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cytlak, U. , Resteu, A. , Bogaert, D. , Kuehn, H. S. , Altmann, T. , Gennery, A. , Jackson, G. et al., Ikaros family zinc finger 1 regulates dendritic cell development and function in humans. Nat. Commun. 2018. 9: 1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cisse, B. , Caton, M. L. , Lehner, M. , Maeda, T. , Scheu, S. , Locksley, R. , Holmberg, D. et al., Transcription factor E2‐2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008. 135: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dontje, W. , Schotte, R. , Cupedo, T. , Nagasawa, M. , Scheeren, F. , Gimeno, R. , Spits, H. et al., Delta‐like1‐induced Notch1 signaling regulates the human plasmacytoid dendritic cell versus T‐cell lineage decision through control of GATA‐3 and Spi‐B. Blood. 2006. 107: 2446–2452. [DOI] [PubMed] [Google Scholar]
- 26. Balan, S. , Arnold‐Schrauf, C. , Abbas, A. , Couespel, N. , Savoret, J. , Imperatore, F. , Villani, A. ‐C. et al., Large‐Scale Human Dendritic Cell Differentiation Revealing Notch‐Dependent Lineage Bifurcation and Heterogeneity. Cell Rep. 2018. 24: 1902–1915.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kirkling, M. E. , Cytlak, U. , Lau, C. M. , Lewis, K. L. , Resteu, A. , Khodadadi‐Jamayran, A. , Siebel, C. W. et al., Notch Signaling Facilitates In Vitro Generation of Cross‐Presenting Classical Dendritic Cells. Cell Rep. 2018. 23: 3658–3672.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sander, J. , Schmidt, S. V. , Cirovic, B. , McGovern, N. , Papantonopoulou, O. , Hardt, A. ‐L. , Aschenbrenner, A. C. et al., Cellular Differentiation of Human Monocytes Is Regulated by Time‐Dependent Interleukin‐4 Signaling and the Transcriptional Regulator NCOR2. Immunity. 2017. 47: 1051–1066.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Durand, M. , Walter, T. , Pirnay, T. , Naessens, T. , Gueguen, P. , Goudot, C. , Lameiras, S. et al., Human lymphoid organ cDC2 and macrophages play complementary roles in T follicular helper responses. J. Exp. Med. 2019. 216: 1561–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Segura, E. , Valladeau‐Guilemond, J. , Donnadieu, M. ‐H. , Sastre‐Garau, X. , Soumelis, V. and Amigorena, S. , Characterization of resident and migratory dendritic cells in human lymph nodes. J. Exp. Med. 2012. 209: 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jongbloed, S. L. , Kassianos, A. J. , McDonald, K. J. , Clark, G. J. , Ju, X. , Angel, C. E. , Chen, C.‐J. J. et al., Human CD141+ (BDCA‐3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross‐presents necrotic cell antigens. J. Exp. Med. 2010. 207: 1247–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu, C. I. , Becker, C. , Metang, P. , Marches, F. , Wang, Y. , Toshiyuki, H. , Banchereau, J. .et al., Human CD141+ dendritic cells induce CD4+ T cells to produce type 2 cytokines. J. Immunol. 2014. 193: 4335–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mittag, D. , Proietto, A. I. , Loudovaris, T. , Mannering, S. I. , Vremec, D. , Shortman, K. , Wu, L. et al., Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J. Immunol. 2011. 186: 6207–6217. [DOI] [PubMed] [Google Scholar]
- 34. Segura, E. , Durand, M. and Amigorena, S. , Similar antigen cross‐presentation capacity and phagocytic functions in all freshly isolated human lymphoid organ‐resident dendritic cells. J. Exp. Med. 2013. 210: 1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crozat, K. , Guiton, R. , Contreras, V. , Feuillet, V. , Dutertre, C. ‐A. , Ventre, E. , Vu Manh, T. ‐ P. et al., The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 2010. 207: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bachem, A. , Güttler, S. , Hartung, E. , Ebstein, F. , Schaefer, M. , Tannert, A. , Salama, A. et al., Superior antigen cross‐presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 2010. 207: 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chiang, M. ‐C. , Tullett, K. M. , Lee, Y. S. , Idris, A. , Ding, Y. , McDonald, K. J. , Kassianos, A. et al., Differential uptake and cross‐presentation of soluble and necrotic cell antigen by human DC subsets. Eur. J. Immunol. 2016. 46: 329–339. [DOI] [PubMed] [Google Scholar]
- 38. Nizzoli, G. , Krietsch, J. , Weick, A. , Steinfelder, S. , Facciotti, F. , Gruarin, P. , Bianco, A. et al., Human CD1c+ dendritic cells secrete high levels of IL‐12 and potently prime cytotoxic T‐cell responses. Blood. 2013. 122: 932–942. [DOI] [PubMed] [Google Scholar]
- 39. Tang‐Huau, T. ‐L. , Gueguen, P. , Goudot, C. , Durand, M. , Bohec, M. , Baulande, S. , Pasquier, B. et al., Human in vivo‐generated monocyte‐derived dendritic cells and macrophages cross‐present antigens through a vacuolar pathway. Nat. Commun. 2018. 9: 2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hubert, M. , Gobbini, E. , Couillault, C. , Manh, T. ‐P. V. , Doffin, A. ‐C. , Berthet, J. , Rodriguez, C. et al., IFN‐III is selectively produced by cDC1 and predicts good clinical outcome in breast cancer. Sci Immunol. 2020. 5. [DOI] [PubMed] [Google Scholar]
- 41. Lauterbach, H. , Bathke, B. , Gilles, S. , Traidl‐Hoffmann, C. , Luber, C. A. , Fejer, G. , Freudenberg, M. A. et al., Mouse CD8alpha+ DCs and human BDCA3+ DCs are major producers of IFN‐lambda in response to poly IC. J. Exp. Med. 2010. 207: 2703–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brown, C. C. , Gudjonson, H. , Pritykin, Y. , Deep, D. , Lavallée, V. ‐P. , Mendoza, A. , Fromme, R. et al., Transcriptional basis of mouse and human dendritic cell heterogeneity. Cell. 2019. 179: 846–863.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klechevsky, E. , Morita, R. , Liu, M. , Cao, Y. , Coquery, S. , Thompson‐Snipes, L. , Briere, F. et al., Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity. 2008. 29: 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schlitzer, A. , McGovern, N. , Teo, P. , Zelante, T. , Atarashi, K. , Low, D. , Ho, A. W. S. et al., IRF4 transcription factor‐dependent CD11b+ dendritic cells in human and mouse control mucosal IL‐17 cytokine responses. Immunity. 2013. 38: 970–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leal Rojas, I. M. , Mok, W. ‐H. , Pearson, F. E. , Minoda, Y. , Kenna, T. J. , Barnard, R. T. , Radford, K. J. et al., Human blood cd1c+ dendritic cells promote th1 and th17 effector function in memory CD4+ T cells. Front. Immunol. 2017. 8: 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Penel‐Sotirakis, K. , Simonazzi, E. , Péguet‐Navarro, J. and Rozières, A. , Differential capacity of human skin dendritic cells to polarize CD4+ T cells into IL‐17, IL‐21 and IL‐22 producing cells. PLoS One. 2012. 7: e45680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Naessens, T. , Morias, Y. , Hamrud, E. , Gehrmann, U. , Budida, R. , Mattsson, J. , Baker, T. et al., Human Lung Conventional Dendritic Cells Orchestrate Lymphoid Neogenesis during Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2020. 202: 535–548. [DOI] [PubMed] [Google Scholar]
- 48. Fenton, T. M. , Kelly, A. , Shuttleworth, E. E. , Smedley, C. , Atakilit, A. , Powrie, F. , Campbell, S. et al., Inflammatory cues enhance TGFβ activation by distinct subsets of human intestinal dendritic cells via integrin αvβ8. Mucosal Immunol. 2017. 10: 624–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cella, M. , Jarrossay, D. , Facchetti, F. , Alebardi, O. , Nakajima, H. , Lanzavecchia, A. , Colonna, M. et al., Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med. 1999. 5: 919–923. [DOI] [PubMed] [Google Scholar]
- 50. Alculumbre, S. G. , Saint‐André, V. , Di Domizio, J. , Vargas, P. , Sirven, P. , Bost, P. , Maurin, M. et al., Diversification of human plasmacytoid predendritic cells in response to a single stimulus. Nat Immunol. 2018. 19: 63–75. [DOI] [PubMed] [Google Scholar]
- 51. Yun, T. J. , Igarashi, S. , Zhao, H. , Perez, O. A. , Pereira, M. R. , Zorn, E. , Shen, Y. et al., Human plasmacytoid dendritic cells mount a distinct antiviral response to virus‐infected cells. Sci Immunol. 2021. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schmidt, B. , Ashlock, B. M. , Foster, H. , Fujimura, S. H. and Levy, J. A. , HIV‐infected cells are major inducers of plasmacytoid dendritic cell interferon production, maturation, and migration. Virology. 2005. 343: 256–266. [DOI] [PubMed] [Google Scholar]
- 53. Takahashi, K. , Asabe, S. , Wieland, S. , Garaigorta, U. , Gastaminza, P. , Isogawa, M. , Chisari, F. V. et al., Plasmacytoid dendritic cells sense hepatitis C virus‐infected cells, produce interferon, and inhibit infection. Proc Natl Acad Sci USA. 2010. 107: 7431–7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tel, J. , Schreibelt, G. , Sittig, S. P. , Mathan, T. S. M. , Buschow, S. I. , Cruz, L. J. , Lambeck, A. J. A. et al., Human plasmacytoid dendritic cells efficiently cross‐present exogenous Ags to CD8+ T cells despite lower Ag uptake than myeloid dendritic cell subsets. Blood. 2013. 121: 459–467. [DOI] [PubMed] [Google Scholar]
- 55. Guillerme, J. ‐B. , Boisgerault, N. , Roulois, D. , Ménager, J. , Combredet, C. , Tangy, F. , Fonteneau, J. ‐ F. et al., Measles virus vaccine‐infected tumor cells induce tumor antigen cross‐presentation by human plasmacytoid dendritic cells. Clin. Cancer Res. 2013. 19: 1147–1158. [DOI] [PubMed] [Google Scholar]
- 56. Aspord, C. , Leloup, C. , Reche, S. and Plumas, J. , pDCs efficiently process synthetic long peptides to induce functional virus‐ and tumour‐specific T‐cell responses. Eur. J. Immunol. 2014. 44: 2880–2892. [DOI] [PubMed] [Google Scholar]
- 57. Hoeffel, G. , Ripoche, A. ‐C. , Matheoud, D. , Nascimbeni, M. , Escriou, N. , Lebon, P. , Heshmati, F. et al., Antigen crosspresentation by human plasmacytoid dendritic cells. Immunity. 2007. 27: 481–492. [DOI] [PubMed] [Google Scholar]
- 58. Di Pucchio, T. , Chatterjee, B. , Smed‐Sörensen, A. , Clayton, S. , Palazzo, A. , Montes, M. , Xue, Y. et al., Direct proteasome‐independent cross‐presentation of viral antigen by plasmacytoid dendritic cells on major histocompatibility complex class I. Nat. Immunol. 2008. 9: 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ito, T. , Yang, M. , Wang, Y. ‐H. , Lande, R. , Gregorio, J. , Perng, O. A. , Qin, X. ‐ F. et al., Plasmacytoid dendritic cells prime IL‐10‐producing T regulatory cells by inducible costimulator ligand. J. Exp. Med. 2007. 204: 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen, W. , Liang, X. , Peterson, A. J. , Munn, D. H. and Blazar, B. R. , The indoleamine 2,3‐dioxygenase pathway is essential for human plasmacytoid dendritic cell‐induced adaptive T regulatory cell generation. J. Immunol. 2008. 181: 5396–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Villani, A. ‐C. , Satija, R. , Reynolds, G. , Sarkizova, S. , Shekhar, K. , Fletcher, J. , Griesbeck, M. et al., Single‐cell RNA‐seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017. 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dutertre, C. ‐A. , Becht, E. , Irac, S. E. , Khalilnezhad, A. , Narang, V. , Khalilnezhad, S. , Ng, P. Y. et al., Single‐Cell Analysis of Human Mononuclear Phagocytes Reveals Subset‐Defining Markers and Identifies Circulating Inflammatory Dendritic Cells. Immunity. 2019. 51: 573–589.e8. [DOI] [PubMed] [Google Scholar]
- 63. Santegoets, S. J. , Duurland, C. L. , Jordanova, E. J. , van Ham, V. J. , Ehsan, I. , Loof, N. M. , Narang, V. et al., CD163+ cytokine‐producing cDC2 stimulate intratumoral type 1 T cell responses in HPV16‐induced oropharyngeal cancer. J. Immunother. Cancer. 2020. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nakamizo, S. , Dutertre, C. ‐A. , Khalilnezhad, A. , Zhang, X. M. , Lim, S. , Lum, J. , Koh, G. et al., Single‐cell analysis of human skin identifies CD14+ type 3 dendritic cells co‐producing IL1B and IL23A in psoriasis. J. Exp. Med. 2021. 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gerhard, G. M. , Bill, R. , Messemaker, M. , Klein, A. M. and Pittet, M. J. , Tumor‐infiltrating dendritic cell states are conserved across solid human cancers. J. Exp. Med. 2021. 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bakdash, G. , Buschow, S. I. , Gorris, M. A. J. , Halilovic, A. , Hato, S. V. , Sköld, A. E. , Schreibelt, G. et al., Expansion of a BDCA1+CD14+ myeloid cell population in melanoma patients may attenuate the efficacy of dendritic cell vaccines. Cancer Res. 2016. 76: 4332–4346. [DOI] [PubMed] [Google Scholar]
- 67. Kvedaraite, E. , Hertwig, L. , Sinha, I. , Ponzetta, A. , Hed Myrberg, I. , Lourda, M. , Dzidic, M. et al., Major alterations in the mononuclear phagocyte landscape associated with COVID‐19 severity. Proc. Natl. Acad. Sci. USA. 2021. 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Winheim, E. , Rinke, L. , Lutz, K. , Reischer, A. , Leutbecher, A. , Wolfram, L. , Rausch, L. et al., Impaired function and delayed regeneration of dendritic cells in COVID‐19. PLoS Pathog. 2021. 17: e1009742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bourdely, P. , Anselmi, G. , Vaivode, K. , Ramos, R. N. , Missolo‐Koussou, Y. , Hidalgo, S. , Tosselo, J. et al., Transcriptional and functional analysis of cd1c+ human dendritic cells identifies a CD163+ subset priming CD8+CD103+ T cells. Immunity. 2020. 53: 335–352.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Girard, M. , Law, J. C. , Edilova, M. I. and Watts, T. H. ., Type I interferons drive the maturation of human DC3s with a distinct costimulatory profile characterized by high GITRL. Sci Immunol. 2020. 5. [DOI] [PubMed] [Google Scholar]
- 71. Coillard, A. and Segura, E. , In vivo Differentiation of Human Monocytes. Front Immunol. 2019. 10: 1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liao, C. ‐T. , Andrews, R. , Wallace, L. E. , Khan, M. W. A. , Kift‐Morgan, A. , Topley, N. , Fraser, D. J. et al., Peritoneal macrophage heterogeneity is associated with different peritoneal dialysis outcomes. Kidney Int. 2017. 91: 1088–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Segura, E. , Touzot, M. , Bohineust, A. , Cappuccio, A. , Chiocchia, G. , Hosmalin, A. , Dalod, M. et al., Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity. 2013. 38: 336–348. [DOI] [PubMed] [Google Scholar]
- 74. Michea, P. , Noël, F. , Zakine, E. , Czerwinska, U. , Sirven, P. , Abouzid, O. , Goudot, C. et al., Adjustment of dendritic cells to the breast‐cancer microenvironment is subset specific. Nat. Immunol. 2018. 19: 885–897. [DOI] [PubMed] [Google Scholar]
- 75. Richter, L. , Landsverk, O. J. B. , Atlasy, N. , Bujko, A. , Yaqub, S. , Horneland, R. , Øyen, O. et al., Transcriptional profiling reveals monocyte‐related macrophages phenotypically resembling DC in human intestine. Mucosal Immunol. 2018. 11: 1512–1523. [DOI] [PubMed] [Google Scholar]
- 76. Watchmaker, P. B. , Lahl, K. , Lee, M. , Baumjohann, D. , Morton, J. , Kim, S. J. , Zeng, R. et al., Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat. Immunol. 2014. 15: 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Patel, V. I. , Booth, J. L. , Duggan, E. S. , Cate, S. , White, V. L. , Hutchings, D. , Kovats, S. et al., Transcriptional classification and functional characterization of human airway macrophage and dendritic cell subsets. J. Immunol. 2017. 198: 1183–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Reynolds, G. , Vegh, P. , Fletcher, J. , Poyner, E. F. M. , Stephenson, E. , Goh, I. , Botting, R. A. et al., Developmental cell programs are co‐opted in inflammatory skin disease. Science. 2021. 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. McGovern, N. , Schlitzer, A. , Gunawan, M. , Jardine, L. , Shin, A. , Poyner, E. , Green, K. et al., Human dermal CD14+ cells are a transient population of monocyte‐derived macrophages. Immunity. 2014. 41: 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Guttman‐Yassky, E. , Lowes, M. A. , Fuentes‐Duculan, J. , Whynot, J. , Novitskaya, I. , Cardinale, I. , Haider, A. et al., Major differences in inflammatory dendritic cells and their products distinguish atopic dermatitis from psoriasis. J. Allergy Clin. Immunol. 2007. 119: 1210–1217. [DOI] [PubMed] [Google Scholar]
- 81. Zaba, L. C. , Fuentes‐Duculan, J. , Eungdamrong, N. J. , Abello, M. V. , Novitskaya, I. , Pierson, K. C. , Gonzalez, J. et al., Psoriasis is characterized by accumulation of immunostimulatory and Th1/Th17 cell‐polarizing myeloid dendritic cells. J. Invest. Dermatol. 2009. 129: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu, Y. , Wang, R. , Jiang, J. , Cao, Z. , Zhai, F. , Sun, W. , Cheng, X. et al., A subset of CD1c+ dendritic cells is increased in patients with tuberculosis and promotes Th17 cell polarization. Tuberculosis (Edinb.). 2018. 113: 189–199. [DOI] [PubMed] [Google Scholar]
- 83. Martin, J. C. , Chang, C. , Boschetti, G. , Ungaro, R. , Giri, M. , Grout, J. A. , Gettler, K. et al., Single‐Cell Analysis of Crohn's Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti‐TNF Therapy. Cell. 2019. 178: 1493–1508.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Laoui, D. , Keirsse, J. , Morias, Y. , Van Overmeire, E. , Geeraerts, X. , Elkrim, Y. , Kiss, M. et al., The tumour microenvironment harbours ontogenically distinct dendritic cell populations with opposing effects on tumour immunity. Nat. Commun. 2016. 7: 13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zilionis, R. , Engblom, C. , Pfirschke, C. , Savova, V. , Zemmour, D. , Saatcioglu, H. D. , Krishnan, I. et al., Single‐Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity. 2019. 50: 1317–1334.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lavin, Y. , Kobayashi, S. , Leader, A. , Amir, E. ‐A. D. , Elefant, N. , Bigenwald, C. , Remark, R. et al., Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single‐Cell Analyses. Cell. 2017. 169: 750–765.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Binnewies, M. , Mujal, A. M. , Pollack, J. L. , Combes, A. J. , Hardison, E. A. , Barry, K. C. , Tsui, J. et al., Unleashing Type‐2 Dendritic Cells to Drive Protective Antitumor CD4+ T Cell Immunity. Cell. 2019. 177: 556–571.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chen, Y. ‐L. , Gomes, T. , Hardman, C. S. , Vieira Braga, F. A. , Gutowska‐Owsiak, D. , Salimi, M. , Gray, N. et al., Re‐evaluation of human BDCA‐2+ DC during acute sterile skin inflammation. J. Exp. Med. 2020. 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jardine, L. , Wiscombe, S. , Reynolds, G. , McDonald, D. , Fuller, A. , Green, K. , Filby, A. et al., Lipopolysaccharide inhalation recruits monocytes and dendritic cell subsets to the alveolar airspace. Nat. Commun. 2019. 10: 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhang, H. , Gregorio, J. D. , Iwahori, T. , Zhang, X. , Choi, O. , Tolentino, L. L. , Prestwood, T. et al., A distinct subset of plasmacytoid dendritic cells induces activation and differentiation of B and T lymphocytes. Proc Natl Acad Sci USA. 2017. 114: 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Leylek, R. , Alcántara‐Hernández, M. , Lanzar, Z. , Lüdtke, A. , Perez, O. A. , Reizis, B. and Idoyaga, J. , Integrated Cross‐Species Analysis Identifies a Conserved Transitional Dendritic Cell Population. Cell Rep. 2019. 29: 3736–3750.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cytlak, U. , Resteu, A. , Pagan, S. , Green, K. , Milne, P. , Maisuria, S. , McDonald, D. et al., Differential IRF8 requirement defines two pathways of dendritic cell development in humans. SSRN Journal. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yin, X. , Chen, S. and Eisenbarth, S. C. , Dendritic cell regulation of T helper cells. Annu. Rev. Immunol. 2021. 39: 759–790. [DOI] [PubMed] [Google Scholar]
- 94. Nutt, S. L. and Chopin, M. , Transcriptional networks driving dendritic cell differentiation and function. Immunity. 2020. 52: 942–956. [DOI] [PubMed] [Google Scholar]
- 95. Locci, M. , Wu, J. E. , Arumemi, F. , Mikulski, Z. , Dahlberg, C. , Miller, A. T. and Crotty, S. , Activin A programs the differentiation of human TFH cells. Nat. Immunol. 2016. 17: 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Heger, L. , Balk, S. , Lühr, J. J. , Heidkamp, G. F. , Lehmann, C. H. K. , Hatscher, L. , Purbojo, A. et al., CLEC10A Is a Specific Marker for Human CD1c+ Dendritic Cells and Enhances Their Toll‐Like Receptor 7/8‐Induced Cytokine Secretion. Front. Immunol. 2018. 9: 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Guilliams, M. , Dutertre, C. ‐A. , Scott, C. L. , McGovern, N. , Sichien, D. , Chakarov, S. , Van Gassen, S. et al., Unsupervised High‐Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity. 2016. 45: 669–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Poulin, L. F. , Reyal, Y. , Uronen‐Hansson, H. , Schraml, B. U. , Sancho, D. , Murphy, K. M. , Håkansson, U. K. et al., DNGR‐1 is a specific and universal marker of mouse and human Batf3‐dependent dendritic cells in lymphoid and nonlymphoid tissues. Blood. 2012. 119: 6052–6062. [DOI] [PubMed] [Google Scholar]
- 99. Galibert, L. , Diemer, G. S. , Liu, Z. , Johnson, R. S. , Smith, J. L. , Walzer, T. , Comeau, M. R. et al., Nectin‐like protein 2 defines a subset of T‐cell zone dendritic cells and is a ligand for class‐I‐restricted T‐cell‐associated molecule. J. Biol. Chem. 2005. 280: 21955–21964. [DOI] [PubMed] [Google Scholar]


