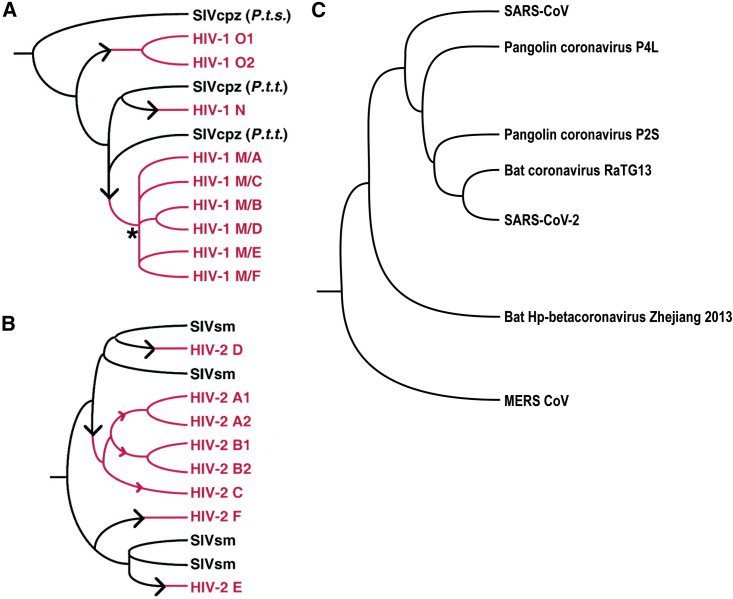
Figure 2.
Zoonotic transmission of SIV to humans and simplified evolution of betacoronaviruses
(A and B) Schematic trees illustrating multiple independent zoonotic transmissions of Simian immunodeficiency virus chimpanzee (SIVcpz) and SIV sootey mangabey (SIVsm) to humans. Branches in black indicate evolution of SIV within its natural hosts, black arrows indicate points of cross-species transmission, and branches in red indicate subsequent evolution within human hosts.
(A) SIVcpz and HIV-1. The three groups of HIV-1 (M, N, and O) are interspersed among SIVcpz strains from P.t. troglodytes (P.t.t.) and P.t. schweinfurthii (P.t.s.). The multiple subtypes of group M derive from a common ancestor indicated by a black asterisk.
(B) SIVsm and HIV-2. The six subtypes of HIV-2 (A–F) are interspersed among SIVsm lineages. Further characterization of SIVsm diversity may reveal that subtypes A, B, and C also arose through separate cross-species transmissions (indicated by the red arrows). For HIV-2, multiple isolates have been found only for subtypes A and B. Reproduced with permission and additional information can be found in Hahn et al.41
(C) Simplified phylogenetic tree showing the evolution of betacoronaviruses. SARS-CoV-1 and SARS-CoV-2 share their closest common ancestor in the sarbecovirus subgenus and have 79% sequence similarity.42 Middle Eastern respiratory syndrome (MERS)-CoV is from a different subgenus (merbecovirus) and shows a more distant relation with SARS-CoV-2, sharing 50% sequence similarity.43 The closest CoV relative of SARS-CoV-2 is bat CoV RaTG13, sharing 96% similarity, and the second closest is pangolin CoV, sharing 90% similarity.42 This suggests that bats and pangolins are intermediate hosts for SARS-CoV-2.