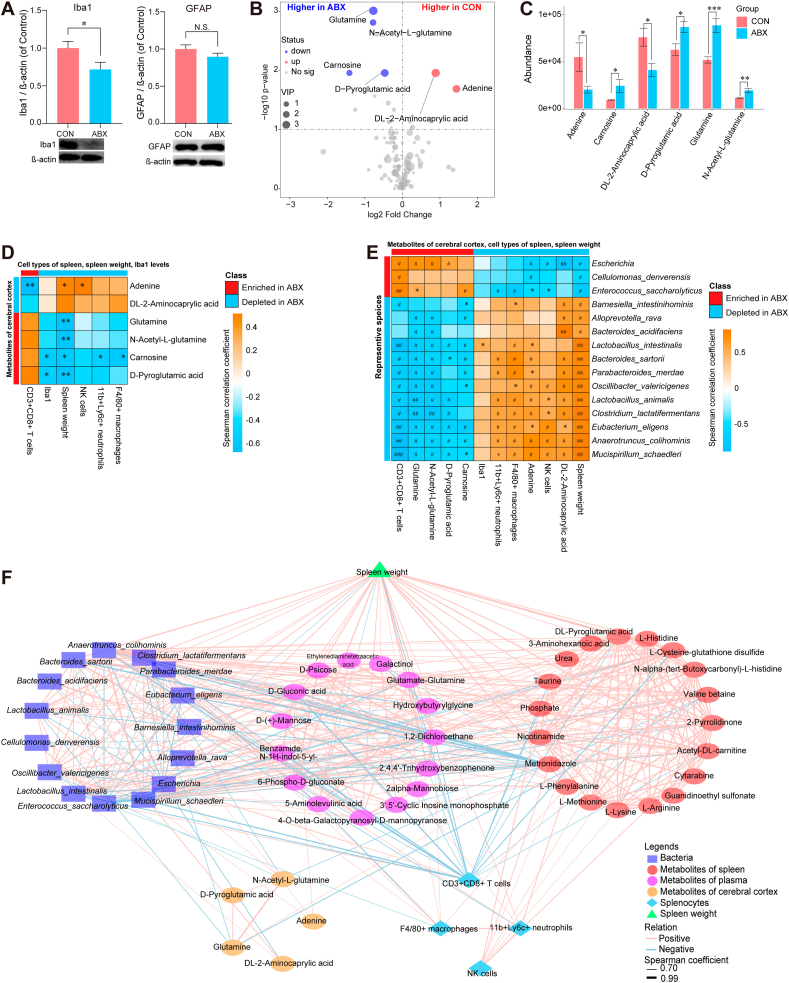
Fig. 6.
Expression of Iba1 and GFAP in the cerebral cortex, altered metabolites in the cerebral cortex, and joint correlation analysis of microbial, multi-location metabolites, and splenic cell populations
A: Expression of Iba1 and GFAP in the cerebral cortex from two groups. B: Volcano plot showing cerebral cortical metabolite profiles between the CON and ABX groups. X-axis indicates the log2 transformed fold change in cerebral cortical metabolite abundance, and y-axis indicates the -log10 transformed P-value using the Wilcox test. Horizontal lines indicate P < 0.05. Red or blue dots indicate differential metabolites between the two groups. C: Differences in abundance of differential metabolites in the cerebral cortex between the two groups (statistical results are presented in Table S6). D: Spearman correlation analysis of differential metabolites in the cerebral cortex with percentage of splenic cell population, Iba1 expression in the cerebral cortex, or spleen weight. E: Spearman correlation analysis of differential species (average of abundance >0.1% in all samples) with differential metabolites in the cerebral cortex, Iba1 level in the cerebral cortex, percentage of splenic cell populations, or spleen weight. F: Joint correlation analysis of spleen weight, percentage of spleen cell population, representative metabolites of different tissues (spleen, plasma, cerebral cortex) and gut microbiota for all mice. Red lines denote positive correlations, blue lines denote negative correlations, and thicker lines represent larger correlation coefficients (Spearman correlation analysis, absolute value of correlation coefficient >0.7, FDR-corrected P < 0.05). Circles indicate metabolites, and different colors indicate originating from different types of tissues. Bacteria are indicated by squares, triangles indicate spleen weights, and diamonds indicate splenic cell populations. Values represent mean ± S.E.M. (CON: n = 8. ABX: n = 9). #P (FDR-corrected) < 0.05, ##P (FDR-corrected) < 0.01, ###P (FDR-corrected) < 0.001. *P < 0.05, **P < 0.01, ***P < 0.001. N.S.: not significant.