Abstract
The number and diversity of drugs in the tuberculosis (TB) drug development process has increased over the years, yet the attrition rate remains very high, signaling the need for continued research in drug discovery. In this study, crude secondary metabolites from marine fungi associated with ascidians collected from Saldanha and False Bays (South Africa) were investigated for antimycobacterial activity. Isolation of fungi was performed by sectioning thin inner-tissues of ascidians and spreading them over potato dextrose agar (PDA). Solid state fermentation of fungal isolates on PDA was then performed for 28 days to allow production of secondary metabolites. Afterwards, PDA cultures were dried and solid-liquid extraction using methanol was performed to extract fungal metabolites. Profiling of metabolites was performed using untargeted liquid chromatography quadrupole time-of-flight tandem mass spectrometry (LC-QTOF-MS/MS). The broth microdilution method was used to determine antimycobacterial activity against Mycobacterium smegmatis mc2155 and Mycobacterium tuberculosis H37Rv, while in silico flexible docking was performed on selected target proteins from M. tuberculosis. A total of 16 ascidians were sampled and 46 fungi were isolated. Only 32 fungal isolates were sequenced, and their sequences submitted to GenBank to obtain accession numbers. Metabolite profiling of 6 selected fungal extracts resulted in the identification of 65 metabolites. The most interesting extract was that of Clonostachys rogersoniana MGK33 which inhibited Mycobacterium smegmatis mc2155 and Mycobacterium tuberculosis H37Rv growth with minimum inhibitory concentrations (MICs) of 0.125 and 0.2 mg/mL, respectively. These results were in accordance with those from in silico molecular docking studies which showed that bionectin F produced by C. rogersoniana MGK33 is a potential inhibitor of M. tuberculosis β-ketoacyl-acyl carrier protein reductase (MabA, PDB ID = 1UZN), with the docking score observed as −11.17 kcal/mol. These findings provided evidence to conclude that metabolites from marine-derived fungi are potential sources of bioactive metabolites with antimycobacterial activity. Even though in silico studies showed that bionectin F is a potent inhibitor of an essential enzyme, MabA, the results should be validated by performing purification of bionectin F from C. rogersoniana MGK33 and in vitro assays against MabA and whole cells (M. tuberculosis).
Keywords: Tuberculosis, Drug-discovery, Marine fungi, MabA, Bionectin F
Tuberculosis; Drug-discovery; Marine fungi; MabA; Bionectin F.
1. Introduction
Tuberculosis (TB) is a serious infectious disease caused by Mycobacterium tuberculosis and is among the top causes of death in HIV-positive individuals [1]. An estimated one quarter of the global population is thought to be infected by the pathogen, yet most of these individuals are not (yet) ill, a condition described as latent TB which is asymptomatic and non-infectious [2]. In 2020, an estimated 5.8 million new TB cases and 1.5 million TB-related deaths were reported worldwide [1].
The effects of both TB and HIV in South Africa are socially and economically catastrophic, this country is among a global group of 30 countries known to have a high TB burden as they collectively contribute 86% of global TB cases [1]. Among these 30 high TB burden countries, South Africa is among the eight countries which contributed two thirds of the global TB cases as follows: “India (26%), China (8.5%), Indonesia (8.4%), the Philippines (6.0%), Pakistan (5.8%), Nigeria (4.6%), Bangladesh (3.6%) and South Africa (3.3%)” [3].
Of great concern is the occurrence and spread of multi-drug resistant-TB (MDR-TB, resistant to isoniazid and rifampicin) and extensively drug resistant TB (XDR-TB, resistant to isoniazid, rifampicin, some of the fluoroquinolones and at least one of the injectable second-line drugs), which are difficult to treat using available drugs [4]. Reported global cases of MDR- and XDR-TB are seemingly increasing every year, yet the rate of discovery of new TB drugs is still somewhat not sufficient to meet an impeding TB pandemic [4]. In 2020, the success rate of MDR- and XDR-TB treatment in South Africa for patients started on second-line TB drugs in 2018 was reported as 65% and 57%, respectively [1].
There is a need for new TB drugs which will increase treatment success rate for both drug susceptible and drug resistant TB strains. Among other side effects of TB drugs, aminoglycosides (including kanamycin, streptomycin and amikacin) are known to cause sensorineural ototoxicity [5], linezolid and ethambutol cause optic neuropathy [6, 7], while isoniazid, pyrazinamide and rifampicin are known to cause hepatotoxicity [8, 9]. Desirable properties for new TB drugs include novel antimycobacterial mechanisms of action, less toxicity, low production costs, limited drug/drug interactions, and high potency [10].
There is abundant evidence in literature that reports secondary metabolites derived from marine fungi as a rich source of bioactive compounds with potential therapeutic uses [11, 12, 13]. Currently, there are 1901 documented marine fungal species [14] and more being discovered annually as interest in bioactive compounds of fungal origin is surging. Marine fungi are commonly isolated as endobionts and/or epibionts of sea animals, plants, algae, corals, and sponges; and less commonly isolated as free-living microbes from sediments [15].
Of particular interest in this study are ascidians, also known as sea-squirts (Phylum: Chordata; Class: Ascidiacea) which are marine invertebrates found all over the world and abundantly in harbors [16]. Ascidians proliferate in nutrient-rich environments and may host a wide diversity of microorganisms in their tissues as it has been shown by the barcoding of their microbiomes [17]. Aspergillus clavatus AS-107 and Aspergillus candidus KMM 4676 are amongst the many examples of fungi isolated from ascidians and were later on shown to produce novel bioactive compounds with antibacterial and cytotoxic activity, respectively [18, 19]. However, studies involving bioprospecting of metabolites from ascidian-associated fungi to discover compounds with bioactivity against M. tuberculosis, are rarely reported.
An interesting example of a marine fungus (not associated with ascidians) that was shown to produce compounds with antimycobacterial activity includes Nigrospora sp., a mangrove endophyte that produced anthraquinone 4-deoxybostrycin [20]. It was observed that 4-deoxybostrycin had activity against M. tuberculosis H37Rv and MDR M. tuberculosis K2903531 in a Kirby-Bauer disk diffusion susceptibility assay (zone of inhibition = 25 mm) [20]. In an in silico study that investigated the antimycobacterial activity of 100 anthraquinones from marine fungi against essential enzymes from M. tuberculosis H37Rv, namely M. tuberculosis β-ketoacyl-acyl carrier protein reductase (MabA) and polyketide synthase 18 (PKS18), averufin produced by Aspergillus sp., was found to be more bioactive than 4-deoxybostrycin [21, 22].
Researchers in this study purposed to investigate the bioactivity of several marine fungi isolated from native and invasive ascidians found along the South African coastline. In vivo antimycobacterial activity of fungal crude extracts were determined against Mycobacterium smegmatis mc2155 and M. tuberculosis H37Rv and minimum inhibitory concentrations were determined. Untargeted liquid chromatography mass spectrometry (LC-MS) was performed, and annotation was done manually to tentatively identify the fungal metabolites. Finally, molecular docking and molecular dynamics simulations were performed to identify putative fungal bioactive metabolites with antimycobacterial potential. This study will immensely contribute to the discovery of new drugs derived from marine fungi bioactive metabolites.
2. Materials and methods
2.1. Collection of ascidians and their identification
Different species of both native and invasive ascidians were collected from Saldanha Bay (33°1′36.06″S, 17° 57′39.55″E) and False Bay (34°11′28.40″S, 18°26′2.96″E) on the 9th and 11th of September 2019 respectively. The two sites serve as marinas for local and international yachts and small boats. Following a modified method outlined by Havenga [23], ascidians were harvested from Perspex® plates that had been tied to a rope and submerged in water for 23 weeks at a depth of 1.5 and 3.0 m. At the time of harvesting, various species of ascidians had settled and grown on the plates, with some species occurring on multiple plates. Ascidians still attached to the plates were then transported the laboratory using 10 L sealable-plastic containers filled with chilled sea water. Upon arriving at the laboratory, the samples were kept at 4 °C until processed within 24 h. Morphological identification of ascidians and their classification was done by marine biologists, Prof. Tamara Bridgett Robinson and Dr. Tainã Gonçalves Loureiro, who considered features such as color, shape, texture, individual and colony size, growth patterns and position of siphons.
2.2. Isolation of fungi from ascidians, DNA sequencing and species identification
Each ascidian sample was detached from the Perspex® plate and then rinsed at least three times with sterile distilled water. Using a sterile scalpel, ascidian samples were then sectioned into small pieces of 0.5 × 0.5 cm and plated onto potato dextrose agar (PDA) in triplicates with four pieces in each plate. Inoculated PDA plates were then incubated at room temperature (23 °C) under constant monitoring for 14 days to promote growth of fungi. After the incubation period, each distinct fungal colony from mixed-culture plates was sub-cultured by streaking on newly prepared PDA and allowed to grow for 7 days. Afterwards, morphological characteristics of fungal colonies were studied to investigate whether the colonies that emerged were pure or mixed cultures. Cultures determined as mixed were once again streaked on fresh media and allowed to incubate for a further 7 days, while pure cultures were re-streaked at least twice and then prepared for long-term storage. Purified fungal cultures were prepared for long-term storage by firstly culturing in potato dextrose broth (PDB) and incubating at room temperature on a rotary shaker for 3 days (or until exponential phase was reached), followed by preparation of 50% culture-glycerol mixtures (v/v) which were stored at -80 °C (Evosafe-Series HF570-86G, Snijders Labs, Laurent Janssensstraat, The Netherlands).
2.3. Fungal DNA isolation, sequencing and phylogenetic analysis
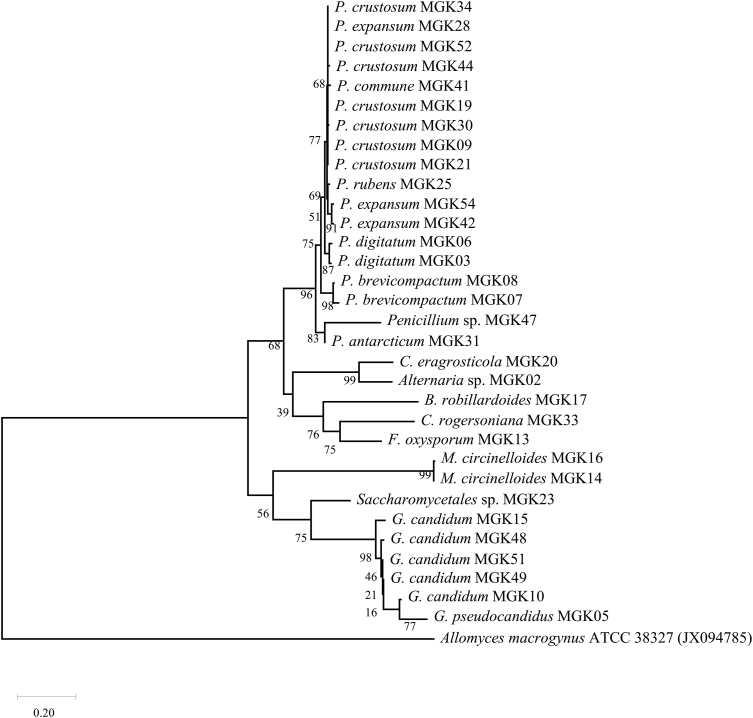
Following a previously described method [24], fungal DNA was extracted and the ribosomal DNA inter-transcribed spacer regions 1 and 2 (ITS 1 and 2) were amplified using the primer pairs ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′). Forward and reverse sequencing was then performed as previously described in Tapfuma et al., [24]. Nucleotide sequences were then visualized and edited using Geneious Prime software (version 2020, http://www.geneious.com/). Using the Nucleotide Basic Local Alignment Search (BLASTN) algorithm on the National Center for Biotechnology Information (NCBI) [25], nucleotide sequences from fungi in this study were compared with sequences from the NCBI database and statistical significance of matches was calculated and assignment of identities was performed. To confirm the assigned identities, maximum likelihood phylogenetic analysis was performed using MEGA X software (version 10, https://www.megasoftware.net/), with Kimura 2-parameter predicted as the best suitable model and Allomyces macrogynus ATCC 38327 used as the outgroup. A total of 1 000 replicate runs were performed, and bootstrap values were calculated. Fungal nucleotide sequences were then submitted to GenBank for curation and accession numbers were assigned (MT738573- MT738604).
2.4. Extraction of crude secondary metabolites from fungi
Fungi were cultured on PDA and incubated at room temperature for 28 days. PDA cultures were then dried in a vented oven blowing a warm stream of air at 30 °C. Dried cultures were then crushed into powder and added to methanol at a ratio of 100 ml of solvent for every 10 g of crushed fungal culture for metabolite extraction [26]. The mixtures were allowed to shake for 24 h before re-extracting the residues with the same amount of solvent 2 more times. The solvent extracts were then filtered through a Whatman No. 1 filter paper and then concentrated using a rotary evaporator at 60 °C under reduced pressure. Concentrated extracts were air-dried under a stream of air at room temperature for 14 days to yield dry and sticky extracts. Concentrated and dried extracts were stored at −80 °C.
2.5. Antimycobacterial activity screening and minimum inhibitory concentrations (MICs) of fungal crude extracts
The preparation of stock solutions of fungal extracts was done by firstly dissolving the extracts in 100% dimethyl sulfoxide (DMSO) and then subsequently diluting them with sterile distilled water to obtain fungal extract solutions with 10% DMSO. Extracts for the actual tests were prepared in Middlebrook 7H9 broth medium containing 0.05% Tween 80, 0.5% glycerol and 10% oleic acid-albumin-dextrose-catalase (OADC) supplement, which was also used in the growth and maintenance of Mycobacterium cultures [27]. Antimycobacterial activity screening was performed at varying concentrations (Table 3) against M. smegmatis mc2155 and M. tuberculosis H37Rv in 96-well microplates for 19 selected fungi which had sufficient extract yields for subsequent experiments. Inoculum was prepared by firstly incubating the starter culture at 37 °C (LOM-400 Series, MRC Laboratory Instruments, Essex, UK) until an optical density (OD600nm) of 0.8–1.0 was reached, followed by adjustment of the culture to OD600nm of 0.01 (Multiskan SkyHigh Microplate Spectrophotometer, Thermo Fisher Scientific, Waltham, MA, USA), subculturing and further incubation until an OD600nm 0.2–0.3 was reached. These cells, at exponential phase were then diluted 100 times in Middlebrook 7H9 broth medium to prepare inoculum for use in screening. Aliquots of 100 μL of fungal extracts and 100 μL of inoculum were added to 96-well microplates and allowed to incubate for 3 days for M. smegmatis mc2155 and 6 days for M. tuberculosis H37Rv. Antimycobacterial activity was determined by addition of 20 μL of 0.015% resazurin dye per well and allowed to incubate for a further 4 h for M. smegmatis mc2155 and 24 h for M. tuberculosis H37Rv [28]. In the presence of viable cells, resazurin (blue) is reduced by viable cells to form resorufin (pink). 96-well microplates were thus visualized after the incubation period and extracts with antimycobacterial activity were noted. Using the broth microdilution assay, minimum inhibitory concentration (MIC) experiments were performed by serially diluting the extracts in 96-well microplates and then following the same procedure outlined for screening. The lowest concentration capable of inhibiting microbial growth after incubation was regarded as the MIC [29]. In all antimycobacterial experiments, isoniazid was used a positive control.
Table 3.
Minimum inhibitory concentration (MIC) values of fungal extracts tested against M. smegmatis mc2155 and M. tuberculosis H37Rv.
| Fungal crude extract | M. smegmatis mc2155 MIC (mg/mL) | M. tuberculosis H37Rv MIC (mg/mL) |
|---|---|---|
| P. digitatum MGK03 | >10 | Not tested |
| P. brevicompactum MGK07 | >5 | Not tested |
| P. brevicompactum MGK08 | 10 | Not tested |
| P. crustosum MGK09 | >10 | Not tested |
| F. oxysporum MGK13 | >1 | >0.33 |
| M. circinelloides MGK14 | >2.50 | >1.67 |
| M. circinelloides MGK16 | >2.50 | >1.67 |
| P. commune MGK19 | >5 | >3.33 |
| C. eragrosticola MGK20 | >2.50 | >1.67 |
| P. rubens MGK25 | >1 | >0.33 |
| P. expansum MGK28 | >2.50 | >1.67 |
| P. crustosum MGK30 | >10 | Not tested |
| P. antarticum MGK31 | >2.50 | >1.67 |
| C. rogersoniana MGK33 | 0.125 | 0.20 |
| P. crustosum MGK34 | >10 | Not tested |
| P. commune MGK41 | >5 | >3.33 |
| P. expansum MGK42 | >5 | >1.67 |
| P. crustosum MGK44 | >5 | >3.3 |
| P. crustosum MGK52 | >5 | >3.33 |
| Isoniazid | 0.031 | <0.31 |
2.6. Metabolite profiling of fungal extracts
Untargeted metabolite profiling of fungal extracts was performed using liquid chromatography quadrupole time-of-flight mass tandem spectrometry (LC-QTOF-MS/MS), following previously described methods [30, 31]. Briefly, 1 mg/mL of each fungal extract dissolved in LC-MS grade methanol was passed through a 0.22 μm nylon membrane syringe filter and then 200 μL of sample was added to LC-MS autosampler vials with inserts and pre-split septate leads. An injection volume of 2 μL per sample was selected for separation of analytes using a Waters Acquity ultraperformance liquid chromatography (UPLC) system, fitted with an Acquity photo-diode array (PDA) detector and an Acquity C18 column with the following dimensions: 130 Å pore size, 1.7 μm particle size, 2.1 mm internal diameter and 100 mm length (Acquity, Waters Corp, Milford, MA, USA). Water acidified with 0.1% formic acid (v/v) (solvent A) and acetonitrile (solvent B) were selected for a gradient elution program set as follows: 0% solvent B between 0-0.5 min; 0–100% solvent B between 0.5-12.00 min; 100% solvent B between 12.00-12.50 min; 100-0% solvent B between 12.50-13.00 min; 0% solvent B between 13.00-15.00 min. Both mobile phases were pumped at a constant flow rate of 0.4 mL/min. The UPLC system was connected to a Waters Synapt G2 QTOF-MS system (Waters Corp, Milford, MA, USA) which acquired data under the following conditions: Centroid mode; positive electro-spray ionization (ESI+) MS/MS mode; nitrogen gas for desolvation at 275 °C at a flow rate of 650 L/h; cone voltage at 15 V; capillary voltage at 3 kV; trap MS collision energy of 6 eV and survey scan of 150–1 500 m/z for MS1 data acquisition; trap MS collision energy of 20 eV (low energy) and 70 eV (high energy), and survey scan of 40–1 500 m/z for MS2 data acquisition. The. raw data files acquired from the Waters LC-QTOF-MS system were then converted to. abf files and further analyzed using MS-Dial software (version 4.24) to identify metabolites [32]. Only [M + H]+ adducts ions with a height of 1000 were considered. Putative compound identifications were assigned to ions which found matches with error ppm <7.0 in the following databases: FungalMet (http://www.fungalmet.org/en/test/), KNapSacK (http://www.knapsackfamily.com/KNApSAcK_Family/) and Dictionary of Natural Products (https://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml).
2.7. Molecular docking
The potential mechanism of anti-mycobacterial activity of the fungal metabolites against mycobacterial proteins was investigated using molecular docking studies. Molecular docking was performed to evaluate the potential association of fungal metabolites with M. tuberculosis β-ketoacyl-acyl carrier protein reductase A (MabA), β-lactamase (Blac), and β-ketoacyl-acyl carrier protein synthase (KasA). The 3D structure of MabA (PDB ID: 1UZN), Blac (PDB ID: 4Q8I) and KasA (PDB ID: 4Q8I) were retrieved from the protein data bank. The 3D structures of fungal compounds were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and ChemSpider (http://www.chemspider.com/). The chemical structures were energy minimized using the OPLS 2005 force field in LigPrep (Maestro 12.7, Schrödinger Release 2021-1, NY, USA, accessed via South African CHPC) [27]. Proteins were prepared using Protein Preparation Wizard (Maestro 12.7, Schrödinger Release 2021-1, NY, USA, accessed via South African CHPC). The protein crystallographic structures were prepared by adding polar hydrogens, removing water molecules, co-crystallized heterogeneous groups, and finally by performing an energy minimization using the OPLS 2005 force field [27, 28]. The Sitemap application (Maestro 12.7, Schrödinger Release 2021-1, NY, USA) was utilized to predict possible binding pockets and the predictions were verified using CASTp (http://sts.bioe.uic.edu/castp/index.html?1uzn) [29]. Using the verified binding site predictions from the Sitemap application, dimensions for outer grid boxes were specified using the size of the Sitemap (1UZN = 56.355 Å3, 4Q8I = 43.208 Å3 and 6P9K = 61.396 Å3) while the inner grid boxes were 10 × 10 × 10 Å with the following grid center of dimensions (x, y, z): 1UZN = 14.896, -18.106, -3.669 Å; for 4Q8I = 28.632, -10.055, 43.208 Å; for 6P9K = 81.502, 49.119, 10.515 Å. The docking was then performed using the Glide application (Maestro 12.7, Schrödinger Release 2021-1, NY, USA, accessed via South African CHPC). The 3D docked poses of the protein-ligand complexes were visualized in Maestro 12.7. The ligands which exhibited the highest docking poses were considered for further analysis with molecular dynamics simulations.
2.8. Molecular dynamics
The best docking pose with the highest binding score from molecular docking was selected for molecular dynamics simulations. The stability of the protein-ligand complex was evaluated through 50 ns (ns) molecular dynamics simulations. Molecular dynamics simulation was performed using applications Desmond module (Schrödinger Release 2021-1, NY, USA, accessed via South African CHPC). The OPLS_2005 force field was used to perform the molecular dynamic simulation. The protein-ligand (1UZN-Bionectin F) complex was explicitly solvated using the TIP3P water model in a boundary condition orthorhombic box shape with boundary dimensions in angstrom units (15 Å × 15 Å × 15 Å) [33]. The system was neutralized by adding 0.15 M counter ions (Na+ and Cl−) to predict the physical properties of a realistic system precisely. The systems were energy minimized and equilibrated in an NPT ensemble at constant temperature and pressure (300 K and 1.01325 bar, respectively) using a default protocol [34]. The equilibrated systems were subjected to a final production step of 50 ns with internal energy recording 1.2 ps (ps) intervals.
3. Results and discussion
3.1. Culture-dependent isolation and identification of fungi associated with ascidians
A total of 16 ascidian samples were collected from Saldanha Bay and False Bay marinas and were identified using their morphological features (Table 1 and Supplementary File 1). In most regions across the world, invasive marine species are introduced by fouling hulls on ships (propeller, rudder, sea chest, etc.), ballast water deposited in new regions and mariculture [35]. In the Cape coastal regions, a great proportion of invasive species originate from the North Atlantic Ocean as a result of shipping patterns (container ships, tour boats, fishing boats, yachts, etc.) and warmer temperatures that prevail [35].
Table 1.
List of sampled ascidians∗, their identities and number of fungi isolated.
| Site | Ascidian code | Characteristics | Common name | Scientific name | Fungal isolates | No. of isolates (%)† |
|---|---|---|---|---|---|---|
| Saldanha Bay (33°1′36.06″S, 17°57′39.55″E) | SB1 | Colonial, native | White ringed ascidian | Botryllus magnicoecus | MGK26 | 1 (2.17%) |
| SB2 | Colonial, native | White ringed ascidian | Botryllus magnicoecus | MGK02, MGK03, MGK06, MGK08, MGK20, MGK31 | 6 (13.04%) | |
| SB3 | Solitary, invasive | Sea vase | Ciona robusta | MGK07, MGK09, MGK10, MGK34, MGK36, MGK51 | 6 (13.04%) | |
| SB4 | Solitary, invasive | Waxy sea squirt | Asterocarpa humilis | - | 0 | |
| SB5 | Colonial, native | White ringed ascidian | Botryllus magnicoecus | MGK25, MGK39, MGK49 | 3 (6.52%) | |
| SB6 | Solitary, invasive | Waxy sea squirt | Asterocarpa humilis | - | 0 | |
| SB7 | Solitary, invasive | Light bulb ascidian | Clavelina lepadiformis | MGK42, MGK48 | 2 (4.35%) | |
| SB8 | Colonial, native | - | Botryllus gregalis | MGK47, MGK53 | 2 (4.35%) | |
| False Bay (34°11′28.40″S, 18°26′2.96″E) | FB1 | Colonial, invasive | Golden star ascidian | Botryllus schlosseri | MGK05, MGK21, MGK35 | 3 (6.52%) |
| FB2 | Solitary, invasive | European sea squirt | Ascidiella aspersa | MGK52, MGK55, MGK56, MGK57 | 4 (8.70%) | |
| FB3 | Solitary, invasive | Light bulb ascidian | Clavelina lepadiformis | MGK14, MGK15, MGK30 | 3 (6.52%) | |
| FB4 | Solitary, invasive | Orange-tipped sea squirt | Corella eumyota | MGK13, MGK16 | 2 (4.35%) | |
| FB5 | Solitary, invasive | European sea squirt | Ascidiella aspersa | MGK17, MGK22, MGK33, MGK41 | 4 (8.70%) | |
| FB6 | Colonial, invasive | Red-rust bryozoan | Watersporia subtorquata | MGK28, MGK46 | 2 (4.35%) | |
| FB7 | Colonial, native | Meandering ascidian | Botryllus meandrus | MGK19, MGK24 | 2 (4.35%) | |
| FB8 | Colonial, native | White ringed ascidian | Botryllus magnicoecus | MGK23, MGK43, MGK44, MGK49, MGK50, MGK54 | 6 (13.04%) |
Photographs of each ascidian sample are available in Supplementary File 1.
Percentage contribution to the overall sum of 46 fungal isolates is shown in brackets () in each row.
Invasive ascidian species accounted for about 75% (6 out of 8) species collected from the False Bay marina, compared to 50% (4 out of 8) of species collected from the Saldanha Bay marina. The higher occurrence of invasive ascidians in False Bay was expected since this marina is a popular tourist site that hosts a higher number of luxury international and local yachts and boats as compared to the Saldanha Bay marina.
Ascidians are filter feeders, their adult bodies are designed as branchial baskets connected to the digestive system while also opening to the exterior to allow for trapping organic matter as the water is filtered through the tissues [36]. Due to this method of feeding, inner tissues of ascidians host a diverse microbiome which is sometimes thought to assist invasive ascidians to adapt to new environments [17], and thus it is possible to isolate numerous microbial symbionts of diverse species. In this study, standard laboratory culture techniques allowed for the isolation of 46 fungi from 94% (15 out of 16) of the collected ascidians. Invasive Ciona robusta (SB3) and native Botryllus magnicoecus (SB1, SB2, SB5 and FB8) species yielded the highest numbers of culturable fungi, while no fungi were isolated from Asterocarpa humilis (SB4 and SB6) (Table 1). Using a culture-independent metagenomics approach could have given more comprehensive insight into the diversity of fungi which associate with ascidians [37, 38], however the limitation of these methods is that they do not allow for in vitro and in vivo bioactivity investigations of compounds produced by the identified fungi. Morphological identification of fungi was performed and identical isolates from the same individual host were eliminated, thus effectively reducing the number of isolates for further experiments to 32.
DNA sequence analysis of the amplified internal transcribed spacer regions 1 (ITS1) and ITS2 of the 32 fungal isolates resulted in the fungi being assigned to 10 genera and 14 species as shown in Table 2 and Figure 1. It was observed that filamentous fungi (Penicillium genus in particular) were predominant and consistent with literature where reports mention that most of the fungi identified from marine environments to date have actually been from the Ascomycota and Basidiomycota phyla [39]. The maximum likelihood phylogenetic tree of the 32 sequences fungal isolates in Figure 1 corroborated the BLAST results from GenBank (partly shown in Table 2) as organisms from the same genus and species formed clads together.
Table 2.
Percentage sequence similarity of fungal isolates and their closest relatives in the GenBank database.
| Fungal isolate | Closest relative in NCBI (accession no.) | Similarity (%) | Assigned identity | GenBank accession no. |
|---|---|---|---|---|
| MGK02 | Alternaria sp. D21 (MH029120) | 99.47 | Alternaria sp. MGK02 | MT738573 |
| MGK03 | Penicillium digitatum CMV010G4 (MK450692) | 100 | P. digitatum MK03 | MT738574 |
| MGK05 | Galactomyces pseudocandidus CBS:11392 (KY103457) | 100 | G. pseudocandidus MGK05 | MT738575 |
| MGK06 | P. digitatum CMV010G4 (MK450692) | 98.98 | P. digitatum MK06 | MT738576 |
| MGK07 | Penicillium brevicompactum (MH047201) | 99.83 | P. brevicompactum MGK07 | MT738577 |
| MGK08 | P. brevicompactum kfb KR704880 | 100 | P. brevicompactum MGK08 | MT738578 |
| MGK09 | Penicillium crustosum DUCC5730 (MT582770) | 99.81 | P. crustosum MGK09 | MT738579 |
| MGK10 | Geotrichum candidum M13 (MN007135.1) | 97.86 | G. candidum MGK10 | MT738580 |
| MGK13 | Fusarium oxysporum A6-2 () | 100 | F. oxysporum MGK13 | MT738581 |
| MGK14 | Mucor circinelloides F1-01 (JN561250) | 99.69 | M. circinelloides MGK14 | MT738582 |
| MGK15 | G. candidum GAD1 (MN638741) | 99.2 | G. candidum MGK15 | MT738583 |
| MGK16 | Mucor circinelloides CMRC 573 (MT603954) | 99.83 | M. circinelloides MGK16 | MT738584 |
| MGK17 | Bartalinia robillardoides CBS 122705 (NR_126145) | 100 | B. robillardoides MGK17 | MT738585 |
| MGK19 | P. crustosum MBRU_F899 (MZ541868) | 99.83 | P. crustosum MGK19 | MT738586 |
| MGK20 | Curvularia eragrosticola BRIP 12538 (NR_158446) | 98.92 | C. eragrosticola MGK20 | MT738587 |
| MGK21 | P. crustosum MBRU_F899 (MZ541868) | 99.63 | P. crustosum MGK21 | MT738588 |
| MGK23 | Saccharomycetales sp. A2 (KC310808) | 99.73 | Saccharomycetales sp. MGK23 | MT738589 |
| MGK25 | Penicillium rubens DTO269E3 (MN413181) | 99.32 | P. rubens MGK25 | MT738590 |
| MGK28 | P. crustosum MBRU_F899 (MZ541868) | 100 | P. expansum MGK28 | MT738591 |
| MGK30 | P. crustosum strain DUCC5730 (MT582770) | 100 | P. crustosum MGK30 | MT738592 |
| MGK31 | Penicillium antarcticum CBS 116939 (KP016829) | 99.83 | P. antarcticum MGK31 | MT738593 |
| MGK33 | Clonostachys rogersoniana YFCC 899 (MW199073) | 99.3 | C. rogersoniana MGK33 | MT738594 |
| MGK34 | P. crustosum MBRU_F899 (MZ541868) | 100 | P. crustosum MGK34 | MT738595 |
| MGK41 | Penicillium commune CSK2_3 (MK460792) | 98.98 | P. commune MGK41 | MT738596 |
| MGK42 | Penicillium expansum 19-14 (MT872092) | 100 | P. expansum MGK42 | MT738597 |
| MGK44 | P. crustosum MBRU_F899 (MZ541868) | 99.49 | P. crustosum MGK44 | MT738598 |
| MGK47 | Penicillium sp. isolate CLE41 MN544011 | 86.65 | Penicillium sp. MGK47 | MT738599 |
| MGK48 | G. candidum strain GC1 KY009607 | 98.66 | G. candidum MGK48 | MT738600 |
| MGK49 | G. candidum DBMY703 (KJ706920) | 99.47 | G. candidum MGK49 | MT738601 |
| MGK51 | G. candidum M13 (MN007135) | 98.93 | G. candidum MGK51 | MT738602 |
| MGK52 | P. crustosum MBRU_F899 (MZ541868) | 100 | P. expansum MGK52 | MT738603 |
| MGK54 | P. expansum 19-14 (MT872092) | 99.66 | P. expansum MGK54 | MT738604 |
Figure 1.
The evolutionary history was inferred by using the maximum likelihood method and Kimura 2-parameter model. The tree with the highest log likelihood (−6061.78) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches, while GenBank accession numbers are also displayed next to the respective sequences. Allomyces macrogynus was used as an outgroup.
Marine fungal species which fall within the Penicillium, Alternaria, Mucor, Fusarium, Galactomyces and Clonostachys genera have been widely reported as associates of ascidians [40, 41, 42], while fungi of the Saccharomycetales, Geotrichum, Galactomyces and Bartalinia genera are reported as associates of ascidians for the first time in this study. Unfortunately, the sample size of different ascidian species utilized in this study was inadequate to conclude on whether the cultivable fungal microbiota isolated in this study associate with specific ascidian species or not.
3.2. Antimycobacterial activity of extracts from fungi associated with ascidians
The 32 fungi listed in Table 2 were cultured on potato dextrose agar (PDA) for 14 days and crude secondary metabolites were extracted using methanol. Some fungi which include Alternaria sp. MGK02, Saccharomycetales sp. MGK23, those from the Galactomyces and Geotrichum genera exhibited poor growth on PDA and thus very low extract-yields were obtained during methanol-solvent extraction. An observed limitation leading to this occurrence was that PDA does not mimic the chemical composition of sea water, the natural environment for the marine fungi in this study, which perhaps negatively impacted the growth of some isolates. In another study, powdered Ciona intestinalis was used as a supplement for growth media with sea water chemical composition and effectively led to the exclusive isolation of Acrostalagmus, Arthopyrenia, Cordyceps and Sporosarcina fungal species from the gut tissues of C. intestinalis [40]. Even though it was understood that different fungal species may have unique nutritional requirements as shown in some studies [40], it was beyond the scope of this study to investigate the response of fungal isolates to different culture media and growth conditions.
A total of 19 fungal cultures exhibited excellent growth on PDA and thus were prioritized for crude metabolite extraction using methanol. The crude extracts were tested in vitro for antimycobacterial activity against M. smegmatis mc2155 and M. tuberculosis H37Rv, and the minimum inhibitory concentrations (MICs) observed are listed in Table 3. According to Awouafack et al., the MIC of crude extracts are considered to be significant if they are below 0.1 mg/mL, moderate if between 0.1-0.625 mg/mL and weak if greater than 0.625 mg/mL [43]. The methanol extract from C. rogersoniana MGK33 therefore exhibited moderate activity against M. smegmatis mc2155 and M. tuberculosis H37Rv with MIC values of 0.125 and 0.20 mg/mL respectively. No data was found for antitubercular activity of C. rogersoniana extracts in literature to make comparisons. Methanol extracts from Penicillium, Fusarium, Galactomyces, Geotrichum and Mucor showed poor inhibitory activity against M. smegmatis mc2155 and M. tuberculosis H37Rv.
Poor activity from extracts of multiple fungi was unsurprising because Mycobacteria are known to possess intrinsic and extrinsic resistance to many antibiotics currently available. Drug-resistance mechanisms utilized by Mycobacteria include the use of a thick, waxy and hydrophobic cell envelope on the pathogen's surface to limit penetration of compounds [44]. Drugs that may possibly make past the cell wall defense system may be met with degrading enzymes such as rifampin inactivating ADP ribosyltransferase, penicillin inactivating β-lactamase, erythromycin ribosome methyltransferase and aminoglycoside acetyltransferase [44, 45, 46]. Protein pumps such as the ATP-binding cassette (ABC) transporters (e.g., Rv1819c, Rv0194, Rv0933, Rv1473, Rv2477c, etc.) are known to facilitate the efflux of β-lactams, macrolides, isoniazid, rifampicin and several other antibiotics [47].
Since some fungal secondary metabolite biosynthetic gene clusters (BGCs) are understood to be silent in standard culture conditions [48], activating these genes in fungal isolate cultures could promote the production of antimycobacterial compounds by the fungi. Some interesting techniques utilized in the stimulation of silent BCGs include the co-culturing of fungi and Mycobacteria to promote cross-talk and activation of silent-pathways in fungi [49, 50]. In one study, researchers observed that there was selective production of aurasperone, desmethylkotanin, TMC-256A1 and malformin C by marine Aspergillus niger when co-cultured with M. smegmatis [51].
More recently, a CRISPR-based transcriptional activation methodology has been developed for filamentous fungi and promises to expand the exploration of silent genes in fungi [52]. Thus said, fungi which did produce bioactive extracts against Mycobacteria may very well have their BGC-induction studies re-explored to assess metabolites which are not observed in standard culture conditions that were used in this study.
3.3. Metabolite profiling of fungal isolates
Six fungal isolates which include F. oxysporum MGK13, M. circinelloides MGK14, C. eragrosticola MGK20, P. antarticum MGK31, C. rogersoniana MGK33 and P. expansum MGK42 were prioritized for metabolite profiling using liquid chromatography quadrupole time-of-flight tandem mass spectrometry (LC-QTOF-MS/MS) in positive mode. MSFinder was used to calculate molecular formulas for the detected [M + H]+ ions. After manually searching in several public and subscription databases, peaks were annotated with predicted compound IDs as shown in Tables 4 and 5. Chemical structures of the identified compounds are available in Supplementary File 2.
Table 4.
Unique metabolites identified from extracts of six selected fungi and their docking scores.
| No. | Proposed ID | Predicted formula | Precursor m/z [M + H]+ | Err ppm | RT (min) | Docking scores (kcal/mol)∗ |
||
|---|---|---|---|---|---|---|---|---|
| 1UZN | 4Q8I | 6P9K | ||||||
| F. oxysporum MGK13 metabolites | ||||||||
| 1 | Dehydrofusaric acid | C10H11NO2 | 178.0860 | 1.44 | 4.69 | −4.80 | −3.71 | −2.62 |
| 2 | Fusarinolic acid | C10H13NO3 | 196.0966 | 1.13 | 4.69 | −6.01 | −4.60 | −5.06 |
| 3 | Fusaric acid | C10H13NO2 | 180.1017 | 1.15 | 5.42 | −5.40 | −3.48 | −3.64 |
| 4 | 3-Dehydrosphinganine | C18H37NO2 | 300.2885 | 4.03 | 11.49 | −2.65 | −2.85 | −4.96 |
| 5 | Beauvericin | C45H57N3O9 | 784.4191 | −2.99 | 11.92 | −4.15 | −1.49 | −2.88 |
| M. circinelloides MGK14 metabolites | ||||||||
| 6 | Pantothenic acid | C9H17NO5 | 220.1183 | −1.60 | 4.69 | −5.49 | −1.68 | −1.92 |
| C. eragrosticola MGK20 metabolites | ||||||||
| 7 | L-Arginine | C6H14N4O2 | 175.1188 | 0.87 | 0.72 | −4.23 | −4.44 | −5.33 |
| 8 | L-Phenylalanine | C9H11NO2 | 166.0859 | 2.15 | 4.57 | −5.34 | −3.77 | −5.61 |
| 9 | N-Acetylsphinganine | C20H41NO3 | 344.3154 | 1.52 | 9.06 | −4.83 | −4.57 | −4.54 |
| 10 | Sphingosine | C18H37NO2 | 300.2911 | −4.66 | 11.40 | −4.50 | −3.63 | −4.20 |
| 11 | Antibiotic BK230 | C57H91N9O14 | 1126.678 | −1.93 | 12.16 | n.d. | n.d. | n.d. |
| P. antarticum MGK31 metabolites | ||||||||
| 12 | Arohynapene A | C18H22O3 | 287.1631 | 3.74 | 0.78 | −6.87 | −4.36 | −5.59 |
| 13 | Chrysogine | C10H10N2O2 | 191.0812 | 1.60 | 5.31 | −6.90 | −5.27 | −7.00 |
| 14 | L-Phe-L-His | C15H18N4O3 | 303.1452 | −0.11 | 5.41 | −7.37 | −4.89 | −4.96 |
| 15 | Cyclo (L-Phe-L-Pro) | C14H16N2O2 | 245.1286 | −0.60 | 5.77 | −6.02 | −4.30 | −5.06 |
| 16 | Penicitrinol P | C16H20O6 | 309.1328 | 1.51 | 6.25 | −5.43 | −4.90 | −4.67 |
| 17 | Trans-Resorcylide | C16H18O5 | 291.1227 | 0.00 | 6.25 | −6.20 | −2.81 | −6.16 |
| 18 | Penexanthone B | C19H20O8 | 377.1257 | −6.93 | 6.42 | −5.32 | n.d. | −4.78 |
| 19 | Cladosporin | C16H20O5 | 293.1385 | −0.51 | 8.17 | −6.19 | −4.80 | −5.29 |
| 20 | Penicisochroman A | C16H18O4 | 275.1273 | 1.80 | 8.17 | −6.44 | −4.53 | −4.97 |
| 21 | Antibiotic TAN 1446A | C17H22O5 | 307.154 | 0.00 | 9.60 | −5.14 | −3.85 | −4.81 |
| 22 | Chrysogeside C | C40H73NO9 | 712.5357 | 0.15 | 11.98 | −5.69 | −5.20 | −5.59 |
| C. rogersoniana MGK33 metabolites | ||||||||
| 23 | Gadusol | C8H12O6 | 205.0693 | 6.69 | 0.67 | −6.22 | −3.65 | −4.67 |
| 24 | Bionectin F | C50H96O8 | 825.7189 | −1.34 | 9.08 | −11.17 | −8.19 | −8.94 |
| 25 | C16 Phytosphingosine | C16H35NO3 | 290.2702 | −4.25 | 7.85 | −2.16 | −1.15 | −3.23 |
| 26 | L-Ile-L-Pro | C11H20N2O3 | 229.1556 | −4.08 | 2.82 | −5.30 | −3.64 | −4.81 |
| 27 | L-Tryptophan | C11H12N2O2 | 205.0977 | −2.67 | 2.82 | −6.46 | −5.73 | −4.55 |
| 28 | L-Tyrosine | C9H11NO3 | 182.0810 | 0.94 | 1.67 | −5.98 | −5.24 | −4.11 |
| P. expansum MGK42 metabolites | ||||||||
| 29 | Dihydrovermistatin | C18H18O6 | 331.1161 | 4.59 | 0.78 | −5.22 | −2.82 | −4.89 |
| 30 | Sativan | C17H18O4 | 287.1266 | 4.14 | 0.78 | −5.75 | −4.24 | −5.16 |
| 31 | Canadensolide | C11H14O4 | 211.0957 | 3.74 | 5.22 | −4.69 | −3.85 | −4.12 |
| No. | Proposed ID | Predicted formula | Precursor m/z [M + H]+ | Err ppm | RT (min) | Docking scores (kcal/mol)∗ |
||
|---|---|---|---|---|---|---|---|---|
| 1UZN | 4Q8I | 6P9K | ||||||
| P. expansum MGK42 metabolites | ||||||||
| 32 | Aurantioclavine | C15H18N2 | 227.1532 | 4.75 | 5.42 | −4.05 | −5.43 | −5.36 |
| 33 | Clavicipitic acid | C16H18N2O2 | 271.1438 | 1.13 | 5.48 | −6.59 | −4.70 | −5.77 |
| 34 | Roquefortine C | C22H23N5O2 | 390.1931 | −1.67 | 5.75 | −9.36 | −3.48 | −4.98 |
| 35 | Roquefortine D | C22H25N5O2 | 392.2077 | 1.03 | 5.76 | −5.31 | −3.66 | −4.48 |
| 36 | (16R)-Hydroxyroquefortine C | C22H23N5O3 | 406.1877 | −0.82 | 6.07 | −5.80 | −4.04 | −4.98 |
| 37 | Cyclo (L-Phe-L-Phe) | C18H18N2O2 | 295.1449 | −2.71 | 6.36 | −6.05 | −3.87 | −6.08 |
| 38 | Dehydrohistidyl-tryptophanyl-diketopiperazine | C17H15N5O2 | 322.1298 | 0.16 | 6.37 | −8.25 | −6.13 | −6.15 |
| 39 | Communesin E | C27H30N4O2 | 443.2454 | −2.82 | 6.55 | −5.60 | −2.91 | −5.37 |
| 40 | Commnesin 470 | C27H34O7 | 471.2389 | 0.36 | 7.06 | −6.25 | −4.02 | −5.17 |
| 41 | 24-Oxocyclocitrinol | C25H34O4 | 399.2537 | −1.79 | 7.13 | −6.03 | −4.69 | −5.36 |
| 42 | Chaetoglobosin E | C32H38N2O5 | 531.2850 | 0.66 | 7.50 | −4.47 | −3.25 | −6.27 |
| 43 | Communesin B | C32H36N4O2 | 509.2880 | 6.10 | 7.50 | −3.80 | −2.66 | −2.58 |
| 44 | Citrinin | C13H14O5 | 251.0912 | 0.80 | 7.65 | −5.80 | −2.23 | −5.02 |
| 45 | Viridicatin | C15H11NO2 | 238.0862 | 0.23 | 7.65 | −6.49 | −4.75 | −4.72 |
| 46 | Chaetoglobosin A | C32H36N2O5 | 529.2690 | 1.32 | 8.28 | −5.73 | −0.76 | −4.03 |
| 47 | Deformylcalbistrin A | C30H40O7 | 513.2877 | −5.90 | 8.43 | −5.32 | −3.38 | −4.49 |
| 48 | Chaetoglobosin F | C32H38N2O5 | 531.2851 | 0.47 | 8.43 | −6.12 | −1.35 | −3.61 |
| 49 | Communesin D | C32H34N4O3 | 523.2708 | −0.83 | 8.49 | −4.75 | −2.80 | −3.94 |
| 50 | Andrastin A | C28H38O7 | 487.2703 | −2.61 | 9.07 | −4.66 | n.d. | −1.45 |
| 51 | Chaetoglobosin D | C32H36N2O5 | 529.2690 | 1.32 | 9.10 | −4.53 | −2.88 | −4.16 |
| 52 | Cytoglobosin D | C32H38N2O4 | 515.2919 | −2.85 | 9.88 | −7.14 | −1.94 | −4.37 |
| 53 | Chaetoglobosin J | C32H36N2O4 | 513.2749 | −0.23 | 10.36 | −5.96 | −2.97 | −4.44 |
| 54 | Chrysogeside A | C40H73NO9 | 712.5344 | 1.98 | 11.51 | −5.99 | −6.41 | −6.71 |
∗Abbreviations represent protein receptors as follows: 1UZN = M. tuberculosis β-ketoacyl-acyl carrier protein reductase A (MabA); 4Q8I = β-lactamase (Blac) and 6P9K = β-ketoacyl-acyl carrier protein synthase (KasA). In the table, n.d. means not docked.
Table 5.
Metabolites common to two or more fungal isolates and their molecular docking scores.
| No. | Proposed ID | Predicted formula | Metabolite occurrence‡ | Precursor m/z [M + H]+ | Err ppm | RT (min) | Docking scores (kcal/mol)∗ |
||
|---|---|---|---|---|---|---|---|---|---|
| 1UZN | 4Q8I | 6P9K | |||||||
| 55 | Choline sulfate | C5H13NO4S | MGK13 | 184.0633 | 1.67 | 0.78 | −4.51 | −3.94 | −3.37 |
| MGK20 | 184.0629 | 4.95 | 0.78 | ||||||
| MGK31 | 184.0633 | 2.76 | 0.78 | ||||||
| 56 | L-Carnitine | C7H15NO3 | MGK13 | 162.1123 | 1.67 | 0.78 | −4.13 | −3.46 | −3.69 |
| MGK14 | 162.1124 | 0.43 | 0.81 | ||||||
| MGK20 | 162.1123 | 1.05 | 0.78 | ||||||
| MGK31 | 162.1127 | −1.43 | 0.78 | ||||||
| MGK42 | 162.1117 | 4.78 | 0.78 | ||||||
| 57 | Adenosine | C10H13N5O4 | MGK20 | 268.1043 | 4.61 | 4.56 | −6.77 | −4.43 | −4.45 |
| MGK33 | 268.1028 | −1.01 | 2.06 | ||||||
| 58 | Cyclo (L-Leu-L-Pro) | C11H18N2O2 | MGK31 | 211.1441 | 0.02 | 5.62 | −5.57 | −4.78 | −5.67 |
| MGK33 | 211.1448 | −3.31 | 3.85 | ||||||
| 59 | Cyclo (L-Pro-L-Val) | C10H16N2O2 | MGK20 | 197.1287 | −1.25 | 4.86 | −5.92 | −4.07 | −5.55 |
| MGK31 | 197.1288 | −1.76 | 5.18 | ||||||
| 60 | Pyridoxamine | C8H12N2O2 | MGK20 | 169.0970 | 0.92 | 4.59 | −5.26 | −5.30 | −6.20 |
| MGK31 | 169.0970 | 0.92 | 4.88 | ||||||
| 61 | Phytosphingosine | C18H39NO3 | MGK13 | 318.3001 | 0.54 | 7.71 | −3.77 | −2.72 | −6.20 |
| MGK14 | 318.3000 | 0.85 | 7.68 | ||||||
| MGK20 | 318.3007 | −1.35 | 7.59 | ||||||
| MGK31 | 318.2993 | 3.06 | 8.63 | ||||||
| MGK42 | 318.2998 | 1.48 | 7.63 | ||||||
| 62 | Sphinganine | C18H39NO2 | MGK13 | 302.3053 | 0.19 | 8.49 | −4.21 | −3.44 | −3.95 |
| MGK14 | 302.3050 | 1.18 | 8.46 | ||||||
| MGK20 | 302.3059 | −1.81 | 8.37 | ||||||
| MGK31 | 302.3046 | 2.51 | 8.60 | ||||||
| MGK42 | 302.3045 | 2.84 | 8.40 | ||||||
| 63 | Citreospirosteroid | C28H44O4 | MGK31 | 445.3283 | 6.61 | 12.12 | −4.70 | -2.43 | −3.02 |
| MGK42 | 445.3310 | 0.53 | 11.96 | ||||||
| 64 | Chrysogeside B | C41H75NO9 | MGK31 | 726.5522 | −1.02 | 12.53 | −6.91 | −6.63 | −6.98 |
| MGK42 | 726.5524 | −1.30 | 12.22 | ||||||
| 65 | Ergosta-4,6,8 (14),22-tetraen-3-one | C28H40O | MGK13 | 393.3153 | −0.27 | 12.23 | −5.13 | −3.30 | −4.80 |
| MGK31 | 393.3164 | −3.08 | 12.49 | ||||||
Abbreviations represent fungal isolates as follows: MGK13 = F. oxysporum MGK13; MGK14 = M. circinelloides MGK14; MGK20 = C. eragrosticola MGK20; MGK31 = P. antarticum MGK31; MGK33 = C. rogersoniana MGK33 and MGK42 = P. expansum MGK42.
Abbreviations represent protein receptors as follows: 1UZN = M. tuberculosis β-ketoacyl-acyl carrier protein reductase A (MabA); 4Q8I = β-lactamase (Blac) and 6P9K = β-ketoacyl-acyl carrier protein synthase (KasA). In the table, n.d. means not docked.
Among the metabolites identified, a total of 55 metabolites were found to be unique (Table 4), and whose occurrence was observed in a single individual fungal isolate in this study. Of these unique metabolites, five were identified from the methanol crude extract of F. oxysporum MGK13 as follows: Dehydrofusaric acid (1), fusarinolic acid (2), fusaric acid (3), 3-dehydrosphinganine (4) and beauvericin (5). Against M. tuberculosis, fusaric acid (2) at a concentration of 60 μM was found to cause 14.9% growth inhibition [53], while beauvericin (5) exhibited a MIC of 0.8–1.6 mg/mL [54].
The weak antimycobacterial activity of fusaric acid (2) and beauvericin (5) help explain the poor activity observed from F. oxysporum MGK13 methanol crude extract in this study. Metabolites from M. circinelloides MGK14 were very challenging to identify as there are a limited number of reports on pure compounds from the Mucor genus. Bioprospecting studies utilizing extracts from this genus are also rare. However, M. circinelloides has been reported to accumulate β-carotene (Vitamin A) [55], which perhaps explains the identification of pantothenic acid (Vitamin B5) (6) using the MS/MS fragmentation data of m/z 220.1183 [M + H]+. M. circinelloides is also known to be an oleaginous fungus capable of producing γ-linolenic [56]. Among the metabolites identified from C. eragrosticola MGK20, two were amino acids, L-arginine (7) and L-phenylalanine (8). These naturally occurring amino acids participate in various metabolic processes in cells, and thus are not expected to possess antimicrobial effect.
Extracts from P. antarticum MGK31 and P. expansum MGK42 had greatest number of database hits (compounds 12–22 and 29–54 respectively) as the Penicillium genus is one of the widely explored natural source of bioactive agents. It was interesting to observe that even though extracts from the Penicilliums evidently harbored chemically diverse metabolites, the extracts showed poor inhibition of Mycobacterial growth in culture. This was not surprising as Mycobacteria are known to possess intrinsic resistance against a wide range of antibiotics as previously mentioned [44].
Metabolite profiling of the methanol extract from C. eragrosticola MGK20 resulted in the identification of gadusol (23), bionectin F (24), C16 phytosphingosine (25), L-Ile-L-Pro (26), L-Tryptophan (27) and L-Tyrosine (28). Among these compounds, the polyprenol polyterpenoid bionectin F (24) was first isolated from Bionectria sp. Y1085 (Clonostachys and Bionectria genera belongs to the Bionectriaceae family) [57]. Unfortunately, the researchers in the previously mentioned study did not investigate any bioactivity of bionectin F. C16 phytosphingosine is a fatty acid which belongs to the family of sphingolipids, compounds abundant in fungi, plants and animal [58]. Başpınar et al., investigated the antimicrobial activity of commercially obtained phytosphingosine and found strong activity against Enterococcus faecalis (MIC = 4 μg/mL) and Bacillus subtilis (MIC = 8 μg/mL), but poor activity against Pseudomonas aeruginosa, Salmonella enterica and Escherichia coli (MIC ≥1024 μg/mL) [59]. Gadusol (23), a metabolite of fungi, algae, bacteria and animals [60], plays a role as an ultra-violet (UV) photo-protectant that absorbs maximally at 268 nm in acidic pHs, and as an antioxidant [61, 62]. Reports on metabolites from C. rogersoniana are very limited and thus the majority of ion peaks could not be annotated with database IDs. Bioactivity-guided fractionation and purification may help to isolate and characterize previously unidentified pure compounds.
3.4. Molecular docking and molecular dynamics simulation of fungal metabolites
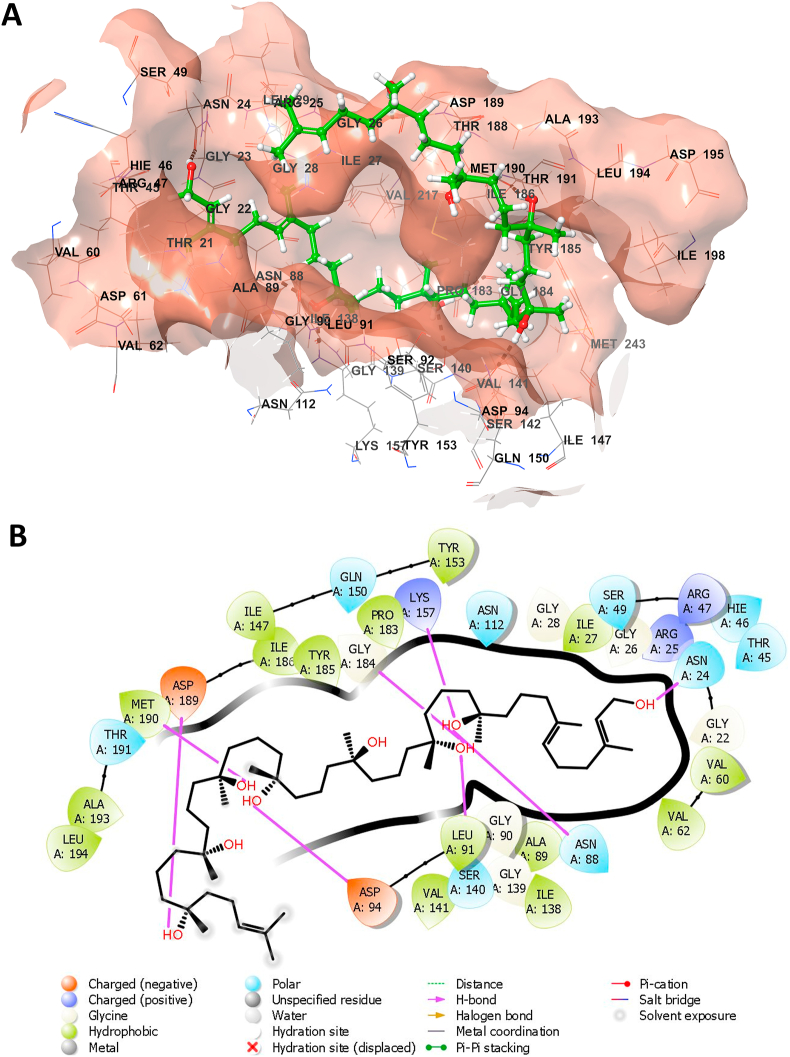
The in silico molecular docking study demonstrated that the investigated ligands interacted with active site residues of M. tuberculosis H37Rv target proteins (1UZN, 4Q8I, and 6P9K) with docking scores presented in Tables 4 and 5. The values of the docking scores revealed that bionectin F has the highest affinity to the selected target proteins (1UZN = −11.17 kcal/mol, 6P9K = −8.94 kcal/mol and 4Q8I = −8.19 kcal/mol). The molecular interactions of bionectin F with residues of the 1UZN and the binding pocket are both depicted in Figures 2A and 2B. The strong interaction of bionectin F with the critical binding cavity of 1UZN was through hydrogen bonding MET A:190, ASP A:189, ASP A:94, LYS A:157, ASN A:88, ASN A:24, ILE A:31 and LYC A:184. Hydrophobic residues in the binding cavity of 1UZN also interacted with bionectin F, thus contributing to the ligand's observed affinity. The 1UZN and bionectin F complex was selected for further analysis in molecular dynamics simulations because of the interestingly high docking affinity.
Figure 2.
Three-dimensional representation of the binding pocket of 1UZN docked with bionectin F (A), and the two-dimensional representation of the molecular interactions observed between bionectin F and the binding site residues of 1UZN docked (B).
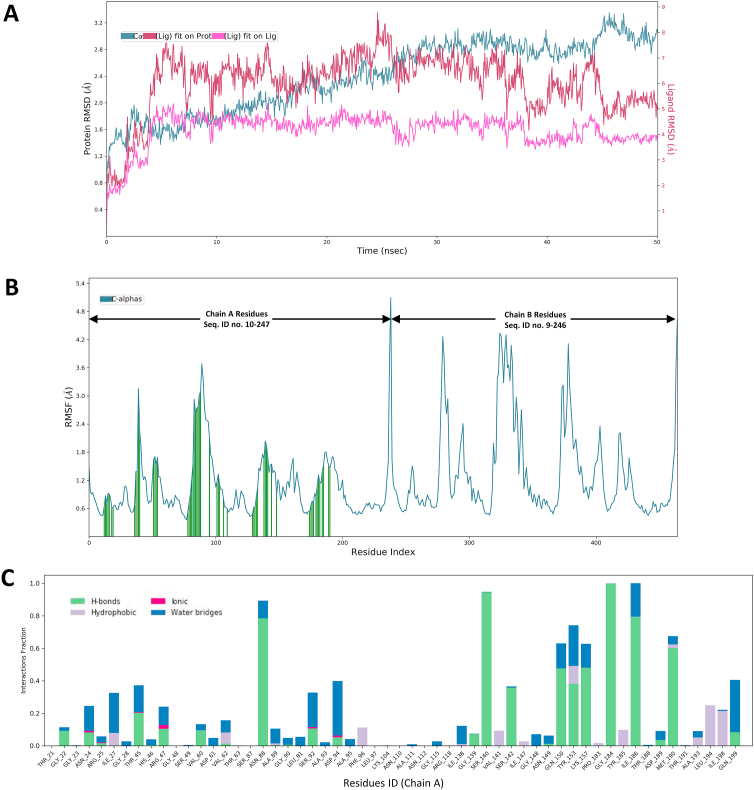
The 1UZN and bionectin F (protein-ligand) complex was subjected to 50 ns molecular dynamics simulations to virtually evaluate in real-time, the structural stability and dynamic behavior of the protein-ligand complex system. The molecular dynamics simulations also determined which amino acids residues of 1UZN interact with the atoms of the bionectin F. The stability of the bionectin F and 1UZN complex was analyzed from the root mean square deviation (RMSD) plot depicted in Figure 3. The RMSD trajectory of the bionectin F and 1UZN complex, sharply augmented from 0 Å to 2.8 Å from the onset until 6 ns, remained constant until around 38 ns and then gradually fluctuated around 2 Å and 2.5 Å. The RMSD of (1UZN) backbone gradually rose from 0 Å to around 2.8 Å from 0 ns until the end of the 50 ns simulation. The RMSD plots show that the bionectin F and 1UZN complex is stable and there were no significant conformational changes in the structure of bionectin F (ligand).
Figure 3.
Molecular dynamics simulation analysis for the 1UZN-bionectin F complex. RSMD plots of the protein-ligand (1UZN and bionectin F) complex, ligand, and the protein back-bone for a 50 ns simulation (A). RMSF plot showing conformational fluctuations of the residues during the simulation, where interaction incidences are depicted by vertical green lines (B). The binding interactions of bionectin F with 1UZN during the simulation (C).
The systemic fluctuations along the 1UZN protein chain residues were analyzed based on the Root Mean Square Fluctuation (RMSF) plot presented in Figure 3A. The results showed low residual fluctuations in residues which interacted with the ligand while larger fluctuations were observed in residues which were not interacting with the ligand. Trajectory analysis of the 0–50 ns simulation showed that bionectin F was bound to the cavity of 1UZN through hydrogen bonds (24 residues), ionic interactions (6 residues), water bridges (36 residues) and hydrophobic interactions (13 residues) along Chain A (Figure 3B). After using CASTp to determine the binding pocket in 1UZN [63], it was noted that A:SER140 forms part of the substrate (β-ketoacyl-acyl carrier protein) binding site, while A:ASN88 and A:ILE186 form part of the binding site of the cofactor (NADPH) (Figure 3C). In a recent study, site-directed mutagenesis on A:SER140 led to complete loss of binding affinity for NADPH which is required by the enzyme to change the conformation the apo-form with a “closed” active site to an open conformation in the holo-form [64, 65].
4. Conclusions
There is growing interest in discovering potent and novel antimycobacterial drugs which will overcome the limitations of presently used TB-drugs. Secondary metabolites from filamentous fungi have been regarded as an inexhaustible reservoir of diverse compounds for consideration in TB-drug discovery programs. In this study, the methanol crude extract from C. rogersoniana MGK33 was found to possess considerable antimicrobial activity against M. smegmatis mc2155 and M. tuberculosis H37Rv, with MICs of 0.125 and 0.2 mg/mL respectively observed. In silico molecular docking studies which of identified metabolites from C. rogersoniana MGK33 revealed that bionectin F is a potential inhibitor of M. tuberculosis β-ketoacyl-acyl carrier protein reductase (MabA), with the docking score observed as -11.17 kcal/mol. The analysis of the molecular dynamics simulations trajectories revealed that bionectin F interacted with the A:SER140, a substrate binding site residue of MabA. The antimycobacterial activity screening results and the molecular docking and molecular dynamics simulations led to the conclusion that bionectin F is a potential inhibitor of MabA, an essential protein in M. tuberculosis. Further work focused on the purification of bionectin F from C. rogersoniana MGK33 and the validating of in silico assays in this study should be performed.
Declarations
Author contribution statement
Vuyo Mavumengwana: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Lucinda Baatjies: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Kudzanai Ian Tapfuma: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Kudakwashe Nyambo: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Rehana Malgas-Enus: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Liezel Smith, Nasiema Allie, Mkhuseli Ngxande, Andre Gareth Loxton, Marshall Keyster:Analyzed: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Francis Adu-Amankwaah: Analyzed and interpreted the data; Wrote the paper.
Funding statement
Vuyo Mavumengwana was supported by National Research Foundation (UID129364).
Mr Kudzanai Ian Tapfuma was supported by Deutscher Akademischer Austauschdienst France (91793243).
Data availability statement
Data associated with this study has been deposited at NCBI GenBank under the accession number MT738573-MT738604.
Declaration of interest's statement
The authors declare no competing interests.
Additional information
Supplementary content related to this article has been published online at https://doi.org/10.1016/j.heliyon.2022.e12406.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.WHO . World Health Organization; Geneva: 2021. Global Tuberculosis Report 2021.https://www.who.int/publications/i/item/9789240037021 (accessed May 30, 2021) [Google Scholar]
- 2.WHO, Tuberculosis (TB) World Health Organ. 2020. https://www.who.int/news-room/fact-sheets/detail/tuberculosis (accessed May 30, 2021)
- 3.TB Statistics, TBFacts.Org. 2022. https://tbfacts.org/tb-statistics/ (accessed October 18, 2022)
- 4.WHO . World Health Organization; Geneva: 2020. Global Tuberculosis Report 2020.https://www.who.int/publications-detail-redirect/9789240013131 (accessed May 30, 2021) [Google Scholar]
- 5.Hong H., Budhathoki C., Farley J.E. Increased risk of aminoglycoside-induced hearing loss in MDR-TB patients with HIV coinfection. Int. J. Tuberc. Lung Dis. Off. J. Int. Union Tuberc. Lung Dis. 2018;22:667–674. doi: 10.5588/ijtld.17.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song W., Si S. The rare ethambutol-induced optic neuropathy. Medicine. 2017;96 doi: 10.1097/MD.0000000000005889. e5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karuppannasamy D., Raghuram A., Sundar D. Linezolid-induced optic neuropathy. Indian J. Ophthalmol. 2014;62:497–500. doi: 10.4103/0301-4738.118451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yee D., Valiquette C., Pelletier M., Parisien I., Rocher I., Menzies D. Incidence of serious side effects from first-line antituberculosis drugs among patients treated for active tuberculosis. Am. J. Respir. Crit. Care Med. 2003;167:1472–1477. doi: 10.1164/rccm.200206-626OC. [DOI] [PubMed] [Google Scholar]
- 9.Jeong I., Park J.-S., Cho Y.-J., Yoon H.I., Song J., Lee C.-T., Lee J.-H. Drug-induced hepatotoxicity of anti-tuberculosis drugs and their serum levels. J. Korean Med. Sci. 2015;30:167–172. doi: 10.3346/jkms.2015.30.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quan D., Nagalingam G., Payne R., Triccas J.A. New tuberculosis drug leads from naturally occurring compounds. Int. J. Infect. Dis. 2017;56:212–220. doi: 10.1016/j.ijid.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 11.Duarte K., Rocha-Santos T.A.P., Freitas A.C., Duarte A.C. Analytical techniques for discovery of bioactive compounds from marine fungi. TrAC, Trends Anal. Chem. 2012;34:97–110. [Google Scholar]
- 12.Deshmukh S.K., Prakash V., Ranjan N. Marine Fungi: a source of potential anticancer compounds. Front. Microbiol. 2018;8 doi: 10.3389/fmicb.2017.02536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C., Tang S., Cao S. Antimicrobial compounds from marine fungi. Phytochem. Rev. 2021;20:85–117. [Google Scholar]
- 14.Marine Fungi, (n.d.). https://www.marinefungi.org/(accessed May 31, 2021).
- 15.El-Bondkly E.A.M., El-Bondkly A.A.M., El-Bondkly A.A.M. Marine endophytic fungal metabolites: a whole new world of pharmaceutical therapy exploration. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watters D.J. Ascidian toxins with potential for drug development. Mar. Drugs. 2018;16:162. doi: 10.3390/md16050162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans J.S., Erwin P.M., Shenkar N., López-Legentil S. Introduced ascidians harbor highly diverse and host-specific symbiotic microbial assemblages. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-11441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Q., Li X.-M., Hu X.-Y., Li X., Chi L.-P., Li H.-L., Wang B.-G. Antibacterial metabolites from Ascidian-derived fungus Aspergillus clavatus AS-107. Phytochem. Lett. 2019;34:30–34. [Google Scholar]
- 19.Yurchenko A.N., Ivanets E.V., Smetanina O.F., Pivkin M.V., Dyshlovoi S.A., von Amsberg G., Afiyatullov Sh.Sh. Metabolites of the marine fungus Aspergillus candidus KMM 4676 associated with a Kuril colonial ascidian. Chem. Nat. Compd. 2017;53:747–749. [Google Scholar]
- 20.Wang C., Wang J., Huang Y., Chen H., Li Y., Zhong L., Chen Y., Chen S., Wang J., Kang J., Peng Y., Yang B., Lin Y., She Z., Lai X. Anti-mycobacterial activity of marine fungus-derived 4-deoxybostrycin and nigrosporin. Molecules. 2013;18:1728–1740. doi: 10.3390/molecules18021728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma A., Islam M.H., Fatima N., Upadhyay T.K., Khan M.K.A., Dwivedi U.N., Sharma R. Elucidation of marine fungi derived anthraquinones as mycobacterial mycolic acid synthesis inhibitors: an in silico approach. Mol. Biol. Rep. 2019;46:1715–1725. doi: 10.1007/s11033-019-04621-0. [DOI] [PubMed] [Google Scholar]
- 22.Hu J., Li Z., Gao J., He H., Dai H., Xia X., Liu C., Zhang L., Song F. New diketopiperazines from a marine-derived fungus strain Aspergillus versicolor MF180151. Mar. Drugs. 2019;17:262. doi: 10.3390/md17050262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Havenga B.S. Stellenbosch University; 2014. Fouling by Non-indigenous marine Species – Impacts on Biodiversity and Mariculture.http://scholar.sun.ac.za/handle/10019.1/86742 (accessed November 23, 2021) [Google Scholar]
- 24.Tapfuma K.I., Uche-Okereafor N., Sebola T.E., Hussan R., Mekuto L., Makatini M.M., Green E., Mavumengwana V. Cytotoxic activity of crude extracts from Datura stramonium’s fungal endophytes against A549 lung carcinoma and UMG87 glioblastoma cell lines and LC-QTOF-MS/MS based metabolite profiling. BMC Complement. Altern. Med. 2019;19:330. doi: 10.1186/s12906-019-2752-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGinnis S., Madden T.L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song R., Wang J., Sun L., Zhang Y., Ren Z., Zhao B., Lu H. The study of metabolites from fermentation culture of Alternaria oxytropis. BMC Microbiol. 2019;19:35. doi: 10.1186/s12866-019-1408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehiro Y., Tomioka H., Pieters J., Tatano Y., Kim H., Iizasa H., Yoshiyama H. Identification of novel mycobacterial inhibitors against mycobacterial protein kinase G. Front. Microbiol. 2018;9:1517. doi: 10.3389/fmicb.2018.01517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elshikh M., Ahmed S., Funston S., Dunlop P., McGaw M., Marchant R., Banat I.M. Resazurin-based 96-well plate microdilution method for the determination of minimum inhibitory concentration of biosurfactants. Biotechnol. Lett. 2016;38:1015. doi: 10.1007/s10529-016-2079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001;48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 30.Tapfuma K.I., Sebola T.E., Uche-Okereafor N., Koopman J., Hussan R., Makatini M.M., Mekuto L., Mavumengwana V. Anticancer activity and metabolite profiling data of Penicillium janthinellum KTMT5. Data Brief. 2020;28:104959. doi: 10.1016/j.dib.2019.104959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magangana T.P., Stander M.A., Masondo N.A., Makunga N.P. Steviol glycoside content and essential oil profiles of Stevia rebaudiana Bertoni in response to NaCl and polyethylene glycol as inducers of salinity and drought stress in vitro, Plant Cell Tissue Organ Cult. PCTOC. 2021;145:1–18. [Google Scholar]
- 32.Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., Ikeda K., Kanazawa M., VanderGheynst J., Fiehn O., Arita M. MS-DIAL: Data Independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods. 2015;12:523. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001;105:9954–9960. [Google Scholar]
- 34.Chinnathambi S., Karthikeyan S., Hanagata N., Shirahata N. Molecular interaction of silicon quantum dot micelles with plasma proteins: hemoglobin and thrombin. RSC Adv. 2019;9:14928–14936. doi: 10.1039/c9ra02829c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson T.B., Peters K., Brooker B. In: Biol. Invasions South Afr. van Wilgen B.W., Measey J., Richardson D.M., Wilson J.R., Zengeya T.A., editors. Springer International Publishing; Cham: 2020. Coastal invasions: the South African context; pp. 229–247. [Google Scholar]
- 36.Gordon T., Roth L., Caicci F., Manni L., Shenkar N. Spawning induction, development and culturing of the solitary ascidian Polycarpa mytiligera, an emerging model for regeneration studies. Front. Zool. 2020;17:19. doi: 10.1186/s12983-020-00365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baeza M., Barahona S., Alcaíno J., Cifuentes V. Amplicon-metagenomic analysis of fungi from Antarctic terrestrial habitats. Front. Microbiol. 2017 doi: 10.3389/fmicb.2017.02235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biller S.J., Berube P.M., Dooley K., Williams M., Satinsky B.M., Hackl T., Hogle S.L., Coe A., Bergauer K., Bouman H.A., Browning T.J., De Corte D., Hassler C., Hulston D., Jacquot J.E., Maas E.W., Reinthaler T., Sintes E., Yokokawa T., Chisholm S.W. Marine microbial metagenomes sampled across space and time. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amend A., Burgaud G., Cunliffe M., Edgcomb V.P., Ettinger C.L., Gutiérrez M.H., Heitman J., Hom E.F.Y., Ianiri G., Jones A.C., Kagami M., Picard K.T., Quandt C.A., Raghukumar S., Riquelme M., Stajich J., Vargas-Muñiz J., Walker A.K., Yarden O., Gladfelter A.S., Garsin D.A. Fungi in the marine environment: open questions and unsolved problems. mBio. 2019;10:e01189–18. doi: 10.1128/mBio.01189-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Utermann C., Echelmeyer V.A., Oppong-Danquah E., Blümel M., Tasdemir D. Diversity, bioactivity profiling and untargeted metabolomics of the cultivable gut microbiota of Ciona intestinalis. Mar. Drugs. 2021;19:6. doi: 10.3390/md19010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.López-Legentil S., Erwin P.M., Turon M., Yarden O. Diversity of fungi isolated from three temperate ascidians. Symbiosis. 2015;66:99–106. [Google Scholar]
- 42.Menezes C.B.A., Bonugli-Santos R.C., Miqueletto P.B., Passarini M.R.Z., Silva C.H.D., Justo M.R., Leal R.R., Fantinatti-Garboggini F., Oliveira V.M., Berlinck R.G.S., Sette L.D. Microbial diversity associated with algae, ascidians and sponges from the north coast of São Paulo state, Brazil. Microbiol. Res. 2010;165:466–482. doi: 10.1016/j.micres.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Awouafack M.D., McGaw L.J., Gottfried S., Mbouangouere R., Tane P., Spiteller M., Eloff J.N. Antimicrobial activity and cytotoxicity of the ethanol extract, fractions and eight compounds isolated from Eriosema robustum (Fabaceae) BMC Complement. Altern. Med. 2013;13:289. doi: 10.1186/1472-6882-13-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gygli S.M., Borrell S., Trauner A., Gagneux S. Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol. Rev. 2017;41:354–373. doi: 10.1093/femsre/fux011. [DOI] [PubMed] [Google Scholar]
- 45.Baysarowich J., Koteva K., Hughes D.W., Ejim L., Griffiths E., Zhang K., Junop M., Wright G.D. Rifamycin antibiotic resistance by ADP-ribosylation: structure and diversity of Arr. Proc. Natl. Acad. Sci. 2008;105:4886–4891. doi: 10.1073/pnas.0711939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanz-García F., Anoz-Carbonell E., Pérez-Herrán E., Martín C., Lucía A., Rodrigues L., Aínsa J.A. Mycobacterial aminoglycoside acetyltransferases: a little of drug resistance, and a lot of other roles. Front. Microbiol. 2019;10:46. doi: 10.3389/fmicb.2019.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Machado D., Lecorche E., Mougari F., Cambau E., Viveiros M. Insights on Mycobacterium leprae efflux pumps and their implications in drug resistance and virulence. Front. Microbiol. 2018;9:3072. doi: 10.3389/fmicb.2018.03072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boruta T. Uncovering the repertoire of fungal secondary metabolites: from Fleming’s laboratory to the International Space Station. Bioengineered. 2018;9:12–16. doi: 10.1080/21655979.2017.1341022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomm H.A., Ucciferri L., Ross A.C. Advances in microbial culturing conditions to activate silent biosynthetic gene clusters for novel metabolite production. J. Ind. Microbiol. Biotechnol. 2019;46:1381–1400. doi: 10.1007/s10295-019-02198-y. [DOI] [PubMed] [Google Scholar]
- 50.Silva K.P.T., Chellamuthu P., Boedicker J.Q. Quantifying the strength of quorum sensing crosstalk within microbial communities. PLoS Comput. Biol. 2017;13:1–16. doi: 10.1371/journal.pcbi.1005809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jomori T., Hara Y., Sasaoka M., Harada K., Setiawan A., Hirata K., Kimishima A., Arai M. Mycobacterium smegmatis alters the production of secondary metabolites by marine-derived Aspergillus niger. J. Nat. Med. 2020;74:76–82. doi: 10.1007/s11418-019-01345-0. [DOI] [PubMed] [Google Scholar]
- 52.Mózsik L., Hoekzema M., de Kok N.A.W., Bovenberg R.A.L., Nygård Y., Driessen A.J.M. CRISPR-based transcriptional activation tool for silent genes in filamentous fungi. Sci. Rep. 2021;11:1118. doi: 10.1038/s41598-020-80864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emani C.S., Williams M.J., Wiid I.J., Baker B., Carolis C. Compounds with potential activity against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.02236-17. e02236-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nilanonta C., Isaka M., Kittakoop P., Trakulnaleamsai S., Tanticharoen M., Thebtaranonth Y. Precursor-directed biosynthesis of beauvericin analogs by the insect pathogenic fungus Paecilomyces tenuipes BCC 1614. Tetrahedron. 2002;58:3355–3360. [Google Scholar]
- 55.Naz T., Nosheen S., Li S., Nazir Y., Mustafa K., Liu Q., Garre V., Song Y. Comparative Analysis of β-carotene production by Mucor circinelloides strains CBS 277.49 and WJ11 under light and dark conditions. Metabolites. 2020;10 doi: 10.3390/metabo10010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang X., Zhao L., Chen H., Chen Y.Q., Chen W., Song Y., Ratledge C. Complete genome sequence of a high lipid-producing strain of Mucor circinelloides WJ11 and comparative genome analysis with a low lipid-producing strain CBS 277.49. PLoS One. 2015;10:1–11. doi: 10.1371/journal.pone.0137543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y.-H., Yang D.-S., Li G.-H., Pu X.-J., Mo M.-H., Zhao P.-J. Antibacterial diketopiperazines from an endophytic fungus Bionectria sp. Y1085. J. Antibiot. 2019;72:752–758. doi: 10.1038/s41429-019-0209-5. [DOI] [PubMed] [Google Scholar]
- 58.Singh A., Del Poeta M. Sphingolipidomics: an important mechanistic tool for studying fungal pathogens. Front. Microbiol. 2016;7:501. doi: 10.3389/fmicb.2016.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Başpinar Y., Kotmakçi M., Öztürk İ. Antimicrobial activity of phytosphingosine nanoemulsions against bacteria and yeasts. Celal Bayar Univ. J. Sci. 2018;14:223–228. [Google Scholar]
- 60.Osborn A.R., Almabruk K.H., Holzwarth G., Asamizu S., LaDu J., Kean K.M., Karplus P.A., Tanguay R.L., Bakalinsky A.T., Mahmud T. De novo synthesis of a sunscreen compound in vertebrates. Elife. 2015;4 doi: 10.7554/eLife.05919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orallo D.E., Lores N.J., Arbeloa E.M., Bertolotti S.G., Churio M.S. Sensitized photo-oxidation of gadusol species mediated by singlet oxygen. J. Photochem. Photobiol., B. 2020;213 doi: 10.1016/j.jphotobiol.2020.112078. [DOI] [PubMed] [Google Scholar]
- 62.Arbeloa E.M., Uez M.J., Bertolotti S.G., Churio M.S. Antioxidant activity of gadusol and occurrence in fish roes from Argentine Sea. Food Chem. 2010;119:586–591. [Google Scholar]
- 63.Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosado L.A., Caceres R.A., de Azevedo W.F., Basso L.A., Santos D.S. Role of Serine140 in the mode of action of Mycobacterium tuberculosis β-ketoacyl-ACP reductase (MabA) BMC Res. Notes. 2012;5:526. doi: 10.1186/1756-0500-5-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Küssau T., Flipo M., Van Wyk N., Viljoen A., Olieric V., Kremer L., Blaise M. Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis β-ketoacyl-acyl carrier protein reductase MabA. Acta Crystallogr. Sect. Struct. Biol. 2018;74:383–393. doi: 10.1107/S2059798318002917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at NCBI GenBank under the accession number MT738573-MT738604.