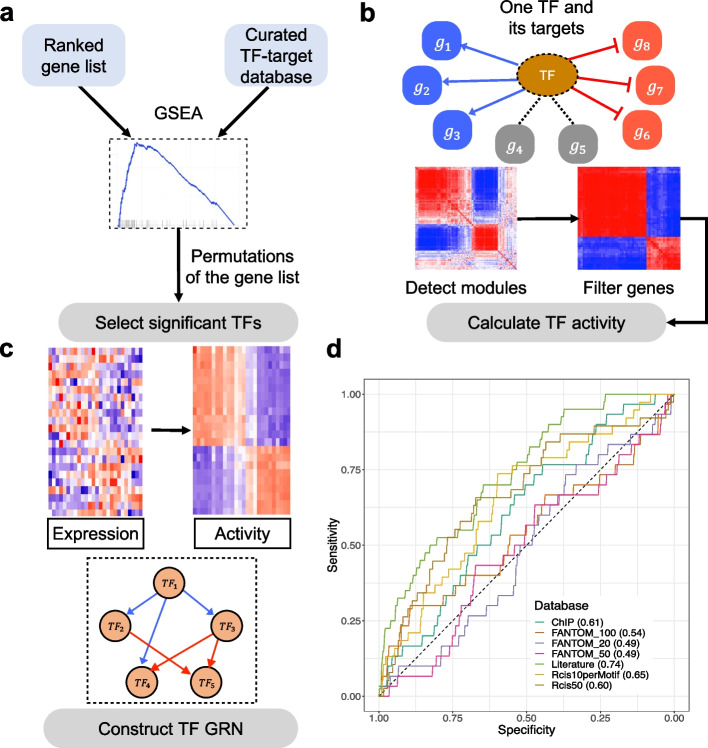
Fig. 1.
Schematics of NetAct. a First, key transcription factors (TFs) are identified using gene set enrichment analysis (GSEA) with a literature-based TF-target database. b Second, the TF activity of an individual sample is inferred from the expression of target genes. From the co-expression and modularity analysis of target genes, we find target genes that are either activated (blue), inhibited (red), or not strongly related to the TF (gray). The activity is defined as the weighted average of target genes activated by the TF minus the weighted average of target genes inhibited by the TF. c Lastly, a TF regulatory network is constructed according to the mutual information of inferred TF activity and literature-based regulatory interactions. d Performance of GSEA for various TF-target gene set databases. The plot shows the sensitivity and specificity with different q-value cutoffs. The gene set databases in the benchmark include the combined literature-based database (D1); FANTOM5-based databases (D2) with 20, 50, and 100 target genes per TF; the combined experimental-based database (D3, ChIP); and RcisTarget databases (D4), one with 10 targets per TF binding motif and another with 50 total number of targets per TF