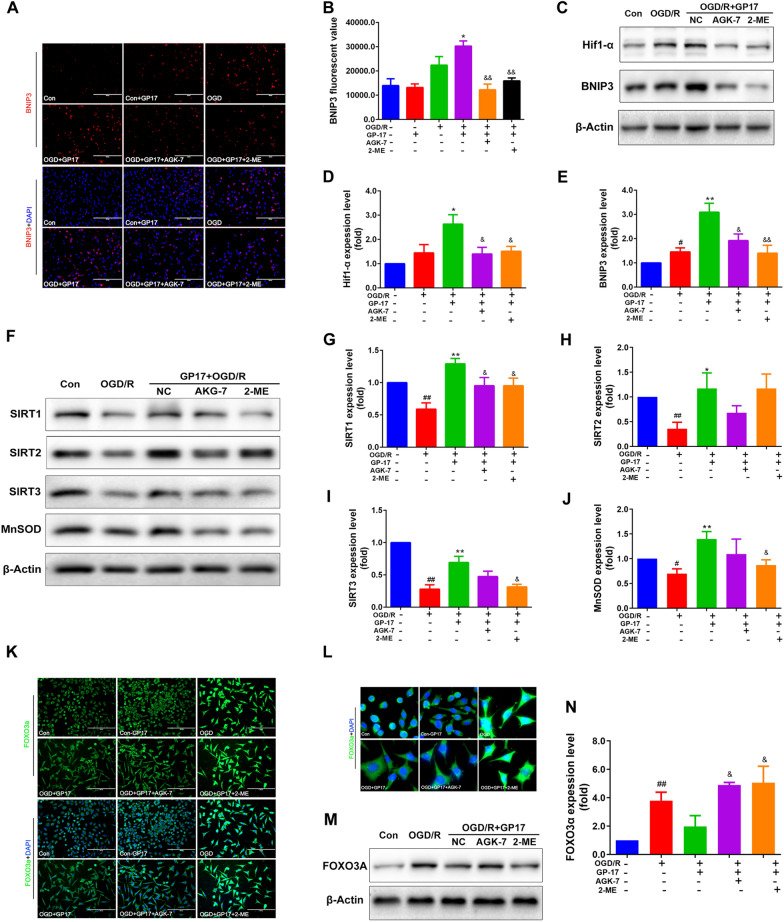
Fig. 6.
Effects of GP17 on the Hif1α-BNIP3 and SIRT1/2/3-FOXO3A signalling pathways in OGD/R-subjected SH-SY5Y cells. These effects were attenuated by the inhibitors AGK-7 and 2-ME. A Double staining of the BNIP3 protein (red) with the nuclei (blue) in OGD/R-subjected SH-SY5Y cells, as located and measured by a fluorescence microscope; scale bar = 200 μm. B Statistical analysis of BNIP3 fluorescence via ImageJ software. C The protein bands of Hif-1α and BNIP3 were examined by western blot analysis in OGD/R-subjected SH-SY5Y cells. D, E The relative expression levels of Hif-1α and BNIP3 in OGD/R-subjected SH-SY5Y cells were quantified and analysed by using Gel-Pro analyser software. The data are presented as the mean values ± SDs (n = 3–6). F. The protein bands of SIRT1/2/3 and MnSOD were examined by western blot analysis in OGD/R-subjected SH-SY5Y cells. G–J The relative expression levels of SIRT1/2/3 and MnSOD in OGD/R-subjected SH-SY5Y cells were quantified and analysed by using Gel-Pro analyser software. The data are presented as the mean values ± SDs (n = 3). K. Double staining of the FOXO3A protein (green) with the nuclei (blue) in OGD/R-subjected SH-SY5Y cells, as located and measured by a fluorescence microscope; scale bar = 100 μm. L Enlarged images of FOXO3A protein. M The relative expression levels of FOXO3A in OGD/R-subjected SH-SY5Y cells were quantified and analysed by using Gel-Pro analyser software. N. The protein bands of FOXO3A were examined by western blot analysis in OGD/R-subjected SH-SY5Y cells. The data presented as Mean values ± SEM. Statistical comparisons were performed as follows: unpaired t test for (B, D, E, G, H, I, J and N); #P < 0.05, ##P < 0.01 versus the control group; *P < 0.05, **P < 0.01 versus the OGD/R model group; &P < 0.05, &&P < 0.01 versus the OGD/R + GP17 group