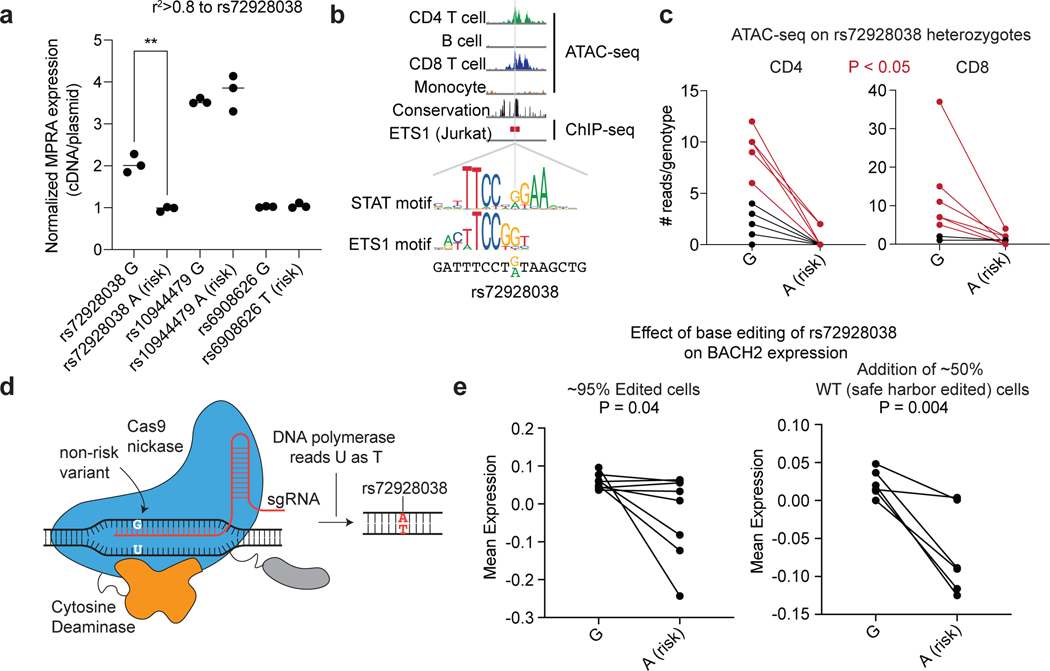
Fig. 4. Base editing of the BACH2 emVar (rs72928038) reduces BACH2 expression.
a) MPRA reporter expression of credible set variant alleles at the BACH2 locus (n=3 independent replicates). b) ATAC-seq profiles in CD4 T cells, B cells, CD8 T cells, and monocytes, vertebrate conservation, and ETS1 ChIP-seq55 at the site of rs72928038 (top). STAT and ETS1 TF motifs at the site of rs72928038 (bottom). c) ATAC-seq reads overlapping rs72928038 in CD4 and CD8 T cells from heterozygous healthy individuals (10 genotyped individuals); 5 of the 10 individuals for CD4 and 6 of the 8 individuals for CD8 (marked red) had a significant difference (at P < 0.05 using a one-sided binomial test, see Supplementary Table 13 for specific P values) in number of reads between reference and alternate alleles. d) Schematic of base editing rs72928038 using the evoCDAmax cytosine base editor. e) PrimeFlow mean expression of BACH2 in cells containing the rs72928038 non-risk (G) and base-edited risk (A) allele with rs72928038 base-edited cells alone (left; 8 independent replicates) and when combined with cells that were edited at a safe harbor locus (right; 6 independent replicates). For (a), ** indicates P = 0.002, according to a two-sided t-test; central tendency is shown as median and all points are plotted to show dispersion. For (e), central tendency is shown as mean and all points are plotted to show dispersion. P values determined by Student’s two-sided t-test (a); one-sided Binomial test (c); Student’s one-sided t-test (e).