FIGURE 4.
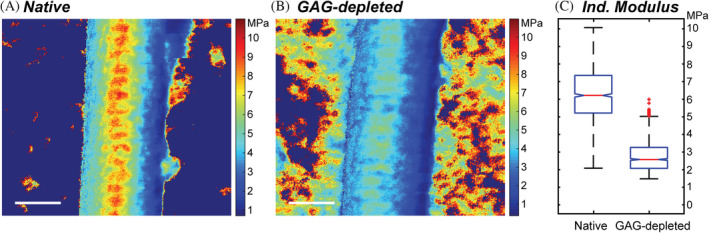
Nanoscale indentation modulus maps of collagen I fibrils in solution. (a,b) Representative 1 × 1 μm fast force maps of individual (a) native and (b) GAG‐depleted collagen fibrils from FFM‐AFM experiments. The color at each pixel shows the local nanoscale indentation moduli (1 pixel/every ~15 nm2). The FFM images are spatial reconstructions of 65,536 discrete force–distance curves that were fitted using standard indentation protocols 53 and subsequently replotted in Cartesian space using in‐house MATLAB code. A large scan size was selected to capture more than 10 D‐bands per image, enabling meaningful comparisons between internal regions, fibrils, and GAG concentration. White scale bars indicate 200 nm. (c) Box and whisker plots compare a subset of indentation moduli data in (a) and (b) for force curves acquired only at the apex down the fibril long axes. In (c), data for five fibrils in each condition (derived from a native or GAG‐depleted tendon taken from the same rat tail) are presented for increased statistical confidence. A one‐way ANOVA analysis confirmed that there is a significant difference between the conditions with a p‐value of <0.01. (Box and whisker plot statistics, Native: IQR = 2.22 MPa, Median = 6.39 MPa, Q1/Q3 = 5.31/7.53 MPa, Outlier cutoffs = [1.98–10.85 MPa], GAG depleted: IQR = 1.13 MPa, Median = 2.64 MPa, Q1/Q3 = 2.13/3.27 MPa Outlier cutoffs = [0.44–4.96 MPa]). ECM, extracellular matrix; FFM, fast force mapping; FFM‐AFM fast force mapping‐atomic force microscopy; GAG, glycosaminoglycans
