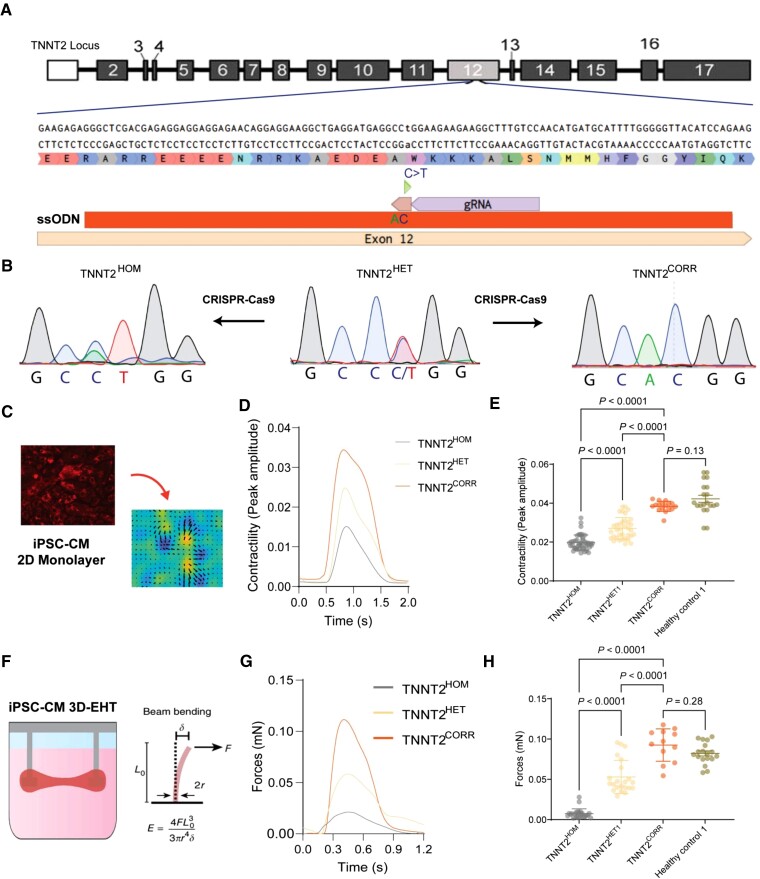
Figure 1.
Generation and validation of the dilated cardiomyopathy in vitro model using induced pluripotent stem cell-cardiomyocytes. (A) Genome editing approaches targeting the TNNT2 c.547C > T mutation. (B) Sanger sequencing of the three isogenic lines: patient heterozygous mutant (TNNT2HET), genome-edited homozygous mutant (TNNT2HOM), and gene-corrected line (TNNT2CORR). (C) High-throughput contractility analysis using vector motion mapping. (D) Representative contractility traces of isogenic induced pluripotent stem cell-cardiomyocytes. (E) Contraction amplitude analyses. Mean ± standard deviation, n = 17–44, three differentiation batches per genotype. (F) Schematics of the 3D-engineered heart tissue contractility analysis. (G) Representative force profile of the isogenic 3D-engineered heart tissues. (H) Quantitation of force generation by the 3D-engineered heart tissues. Mean ± standard deviation. n = 12–24 from four differentiation batches.