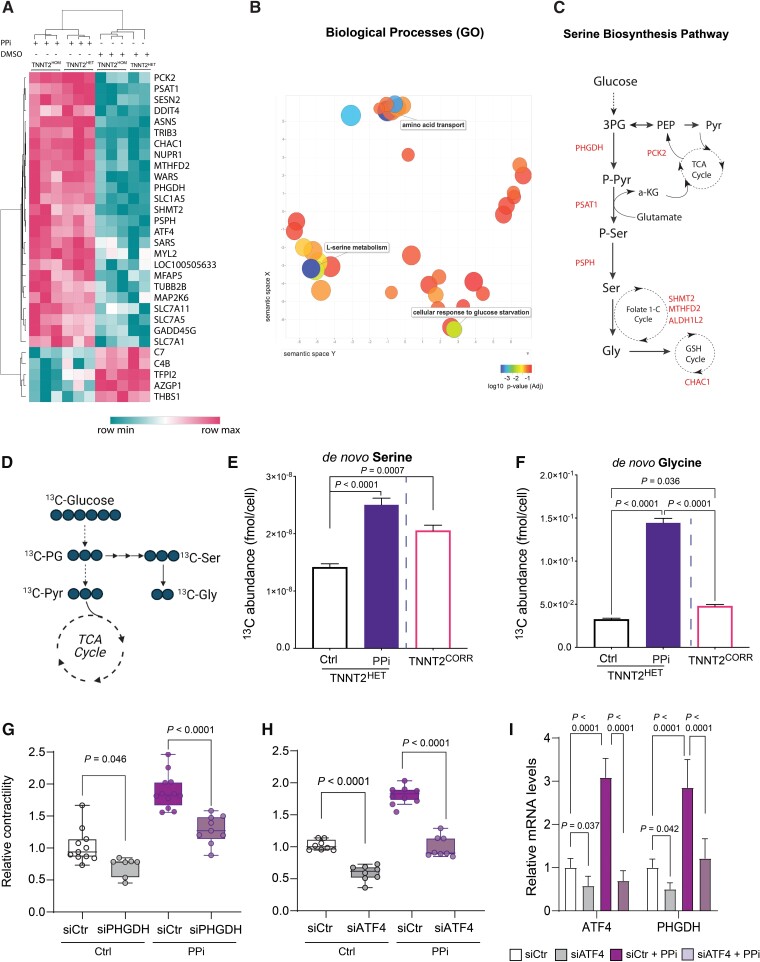
Figure 3.
PPi activates the de novo serine biosynthesis pathway. (A) Heat map illustrating levels of expression of the top 30 differentially expressed genes in TNNT2HOM and TNNT2HET induced pluripotent stem cell-cardiomyocytes after PPi treatment vs. vehicle control (DMSO) (False discovery rate [FDR] < 0.05). (B) Gene Ontology biological processes enrichment analysis of the upregulated transcripts. (C) Graphical representation of enzymes and metabolites of the de novo serine biosynthesis pathway and their integration in cellular metabolism. Genes significantly upregulated in dilated cardiomyopathy-induced pluripotent stem cell-cardiomyocytes upon PPi treatment are indicated throughout the pathway. (D) Schematic showing expected labelling from carbon flow from glucose to serine and glycine when labelled with [13C6]-glucose. (E and F) Abundance of [13C6]-glucose-derived serine and glycine in TNNT2HET induced pluripotent stem cell-cardiomyocytes cultured with [13C6]-glucose for 72 h post-treatment with vehicle control (Ctrl) or PPi. Vehicle control-treated TNNT2CORR induced pluripotent stem cell-cardiomyocytes are also shown for comparison. Data represent mean ± standard deviation, n = 9–18 replicates per condition, two independent labelling experiments. (G) Relative contractility of siRNA control- or siPHGDH-transfected TNNT2HET induced pluripotent stem cell-cardiomyocytes treated with PPi or vehicle control (Ctrl). Box-and-whisker plots show the minimum, the 25th percentile, the median, the 75th percentile, and the maximum. n = 9–12. (H) Relative contractility of siRNA control- or siATF4-transfected TNNT2HET induced pluripotent stem cell-cardiomyocytes treated with PPi or vehicle control (Ctrl). Box-and-whisker plots show the minimum, the 25th percentile, the median, the 75th percentile, and the maximum. n = 9–12. (I) Relative expression of ATF4 and PHGDH in siRNA control- or siATF4-transfected TNNT2HET induced pluripotent stem cell-cardiomyocytes treated with PPi or vehicle control (Ctrl). Mean ± standard deviation, n = 6–12. 1-C, one-carbon; 3PG, 3-phosphoglycerate; a-KG, alpha-ketoglutarate; ALDH1L2, aldehyde dehydrogenase 1 family member L2; CHAC1, glutathione-specific gamma-glutamylcyclotransferase 1; Gly, glycine; MTHFD2, methylenetetrahydrofolate dehydrogenase 2; PCK2, phosphoenolpyruvate carboxykinase 2, mitochondrial; PEP, phosphoenolpyruvate; PHGDH, phosphoglycerate dehydrogenase; PSAT1, phosphoserine aminotransferase 1; PSPH, phosphoserine phosphatase; Pyr, pyruvate; Ser, serine; SHMT2, serine hydroxymethyltransferase 2; TCA, tricarboxylic acid.