Figure 5.
Isolation of a monoclonal NAb that targets a previously undefined site of vulnerability on GPC
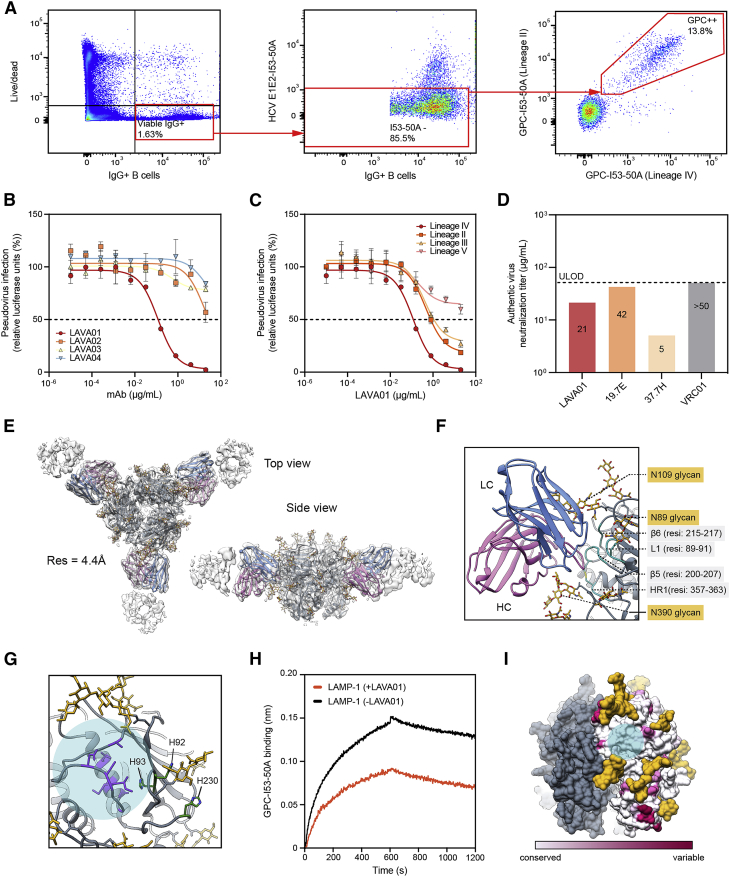
(A) Representative gating strategy for the isolation of GPC-specific B cells. The lymphocyte population was selected, and doublets were excluded (not shown). From live IgG+ B cells (left), cells were selected that showed low reactivity to I53-50A (middle) after which the double-positive GPC-specific B cells were sorted (right).
(B) Pseudovirus neutralization of lineage IV (Josiah) by LAVA01-LAVA04. The dotted line indicates 50% neutralization. Shown are the mean and SEM of two technical replicates.
(C) Pseudovirus neutralization of lineages II, III, IV, and V by LAVA01. The dotted line indicates 50% neutralization. Shown are the mean and SEM of two technical replicates.
(D) Endpoint neutralization titers against authentic LASV (lineage IV) by LAVA01, 19.7E, 37.7H, and VRC01 (HIV-1 Env-specific mAb; negative control). The dotted line indicates the upper limit of detection.
(E) Reconstructed 3D map (transparent white surface) and relaxed model (GPC—gray, glycans—golden, LAVA01 heavy chain—purple, LAVA01 light chain—blue) shown in top and side view.
(F) Peptide (turquoise) and glycan (golden) epitope components for LAVA01 antibody.
(G) Overlay of the LAVA01 footprint (transparent light blue circle) with the histidine triad (green) and proposed LAMP-1 receptor-binding site residues determined by mutagenesis (purple).
(H) Representative sensorgram from BLI experiments showing the binding of recombinant LAMP-1 ectodomain to GPC-I53-50A, in the presence (+LAVA01) or absence (−LAVA01) of LAVA01.
(I) Sequence conservation of GPC with the LAVA01 footprint shown (transparent light blue circle).