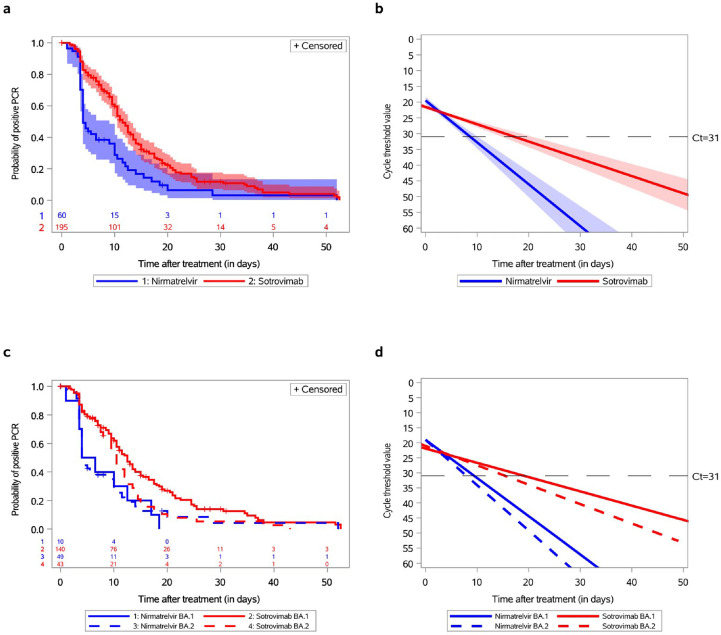
Fig. 1.
Time to negative conversion of nasopharyngeal SARS-CoV-2 PCR and changes in cycle threshold (Ct) values of gene N according to treatment as well as Omicron BA.1 and BA.2 sub-lineages. (a) The survival probability plot for the event of negative PCR conversion amongst 60 Omicron-infected patients treated with nirmatrelvir and 195 Omicron-infected patients treated with sotrovimab (not adjusted on variables at baseline). The p for the log-rank test (p < 0.0001) indicates strong evidence of a significant difference between the survival curves. (b) Mixed model for Ct of gene N for 60 Omicron-infected patients treated with nirmatrelvir and 195 Omicron-infected patients treated with sotrovimab (not adjusted on variables at baseline). The p for the slope was <0.0001. After adjustment for age class and immunosuppression status at baseline, the p for the slope remained <0.0001. (c) The survival probability plots for the event of negative PCR conversion for 10 BA.1-infected patients treated with nirmatrelvir vs. 140 BA.1-infected patients treated with sotrovimab (the p for the log-rank test was 0.01) and for 49 BA.2-infected patients treated with nirmatrelvir vs. 43 BA.2-infected patients treated with sotrovimab (the p for the log-rank test was 0.1). The p was 0.06 for the comparison of BA.1- vs. BA.2-infected patients treated with sotrovimab and 0.84 for the comparison of BA.1- vs. BA.2-infected patients treated with nirmatrelvir. (d) Mixed model for Ct of gene N for 10 BA.1-infected and 49 BA.2-infected patients treated with nirmatrelvir and 140 BA.1-infected and 43 BA.2-infected patients treated with sotrovimab. The p for the slope was 0.003 for BA.1-infected patients and 0.001 for BA.2-infected patients. After adjustment for age class and immunosuppression status at baseline, the p for the slope remained at 0.003 and 0.001 respectively. Ct, cycle threshold.