Abstract
A structural protein of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), nucleocapsid (N) protein is phosphorylated by glycogen synthase kinase (GSK)-3 on the serine/arginine (SR) rich motif located in disordered regions. Although phosphorylation by GSK-3β constitutes a critical event for viral replication, the molecular mechanism underlying N phosphorylation is not well understood. In this study, we found the putative alpha-helix L/FxxxL/AxxRL motif known as the GSK-3 interacting domain (GID), found in many endogenous GSK-3β binding proteins, such as Axins, FRATs, WWOX, and GSKIP. Indeed, N interacts with GSK-3β similarly to Axin, and Leu to Glu substitution of the GID abolished the interaction, with loss of N phosphorylation. The N phosphorylation is also required for its structural loading in a virus-like particle (VLP). Compared to other coronaviruses, N of Sarbecovirus lineage including bat RaTG13 harbors a CDK1-primed phosphorylation site and Gly-rich linker for enhanced phosphorylation by GSK-3β. Furthermore, we found that the S202R mutant found in Delta and R203K/G204R mutant found in the Omicron variant allow increased abundance and hyper-phosphorylation of N. Our observations suggest that GID and mutations for increased phosphorylation in N may have contributed to the evolution of variants.
Keywords: Axin, Delta and Omicron variants, glycogen synthase kinase-3, nucleocapsid, phosphorylation, severe acute respiratory syndrome coronavirus 2
INTRODUCTION
The nucleocapsid (N) protein is the most abundantly expressed structural protein during viral replication of coronavirus (Shah and Woo, 2021; Shan et al., 2021). In addition to its structural role in the ribonucleoprotein complex in virion, the N protein plays key roles in viral RNA and protein synthesis, packaging, and envelope formation (Chang et al., 2014; de Haan and Rottier, 2005). Although the spike protein is only used as an immunogen in current vaccines, serological antibodies against N protein can be used for detection of early and previous infection (Krutikov et al., 2021; Tan et al., 2004). The highly basic N protein of coronavirus consists of about 400 amino acids (~50 kDa) with three distinct domains. Highly conserved N-terminal and C-terminal domains in coronavirus play diverse roles in multimerization and RNA binding (Chang et al., 2014; Shah and Woo, 2021). The central disordered region harbors the Ser/Arg (SR)-rich motif, which is also highly conserved in other coronaviruses, such as OC43, HKU1, and MERS-CoV (Middle East respiratory syndrome-related coronavirus). Interestingly, the SR-rich motif of coronavirus is phosphorylated by glycogen synthase kinase (GSK)-3, and GSK-3 inhibitors suppress viral replication in Vero cells, indicating that GSK-3 mediated N phophosphorylation is a rate-limiting step for viral replication (Peng et al., 2008; Wu et al., 2009). GSK-3 inhibition also selectively reduces genomic RNA and long subgenomic mRNA (Wu et al., 2014), and a clinical trial with a GSK-3 inhibitor, lithium, yielded reduced risk of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) (Liu et al., 2021). Crucially, a recent study revealed that N mutations around the SR-rich motif found in SARS-CoV-2 variants allow significantly increased viral replication compared with ancestral Wuhan Hu-1 N protein (Syed et al., 2021). These results indicate the importance of N phosphorylation by GSK-3 during viral replication and mutational evolution of coronavirus although the underlying molecular mechanism is unclear.
GSK-3 is an endogenously abundant kinase which plays a key role in many signaling pathways, including Wnt signaling. In the Wnt pathway, GSK-3 is recruited to a multi-protein complex with adenomatous polyposis coli (APC) and scaffolding protein Axin (Cohen and Frame, 2001; Doble and Woodgett, 2003). Although Axin binds to the C-terminal of GSK-3 apart from its catalytic site, Axin scaffold assembling with APC and β-catenin decreases GSK-3 kinase activity during the regulation of canonical Wnt activity (Hedgepeth et al., 1999). Crystallography of GSK-3 and Axin peptide reveals that hydrophobic helical ridges formed by Axin residues of Phe388, Leu392, and Leu396 in GID pack into the helical groove of the 285-299 loop in GSK-3, at a distance from the ATP binding site (Dajani et al., 2003). This interacting domain is also found in other GSK-3 interacting proteins, such as FRATs, GSK-3 interacting protein (GSKIP), and WWOX (Howng et al., 2010; Wang et al., 2012). The GID in many proteins provides diverse functions for GSK-3 and its substrate. For example, a multi-protein complex of APC and GSK-3 enhances β-catenin phosphorylation and Axin-GSK3 binding provides nuclear export of GSK-3 stabilizing, epithelial-mesenchymal transition (EMT)-inducer Snail in cancer (Dajani et al., 2003; Yook et al., 2006). The typical amino acids in helical GID consist of L/FxxxL/AxxRL (Fig. 1A). In this study, we found GID located next to the SR-rich domain of N protein in SARS-CoV-2 (hereafter N), critical for N protein phosphorylation, suggesting that interaction between N and GSK-3 plays a critical role in viral replication and evolution of SARS-CoV-2. Compared to N protein in other non-pathogenic coronaviruses, N protein of SARS-CoV-2 also harbors a CDK1 phosphorylation site and a flexible linker between GID and the SR-rich region, allowing enhanced phosphorylation of N. Further, we found that N mutations in Delta and Omicron variants provide enhanced phosphorylation and increased abundance of N.
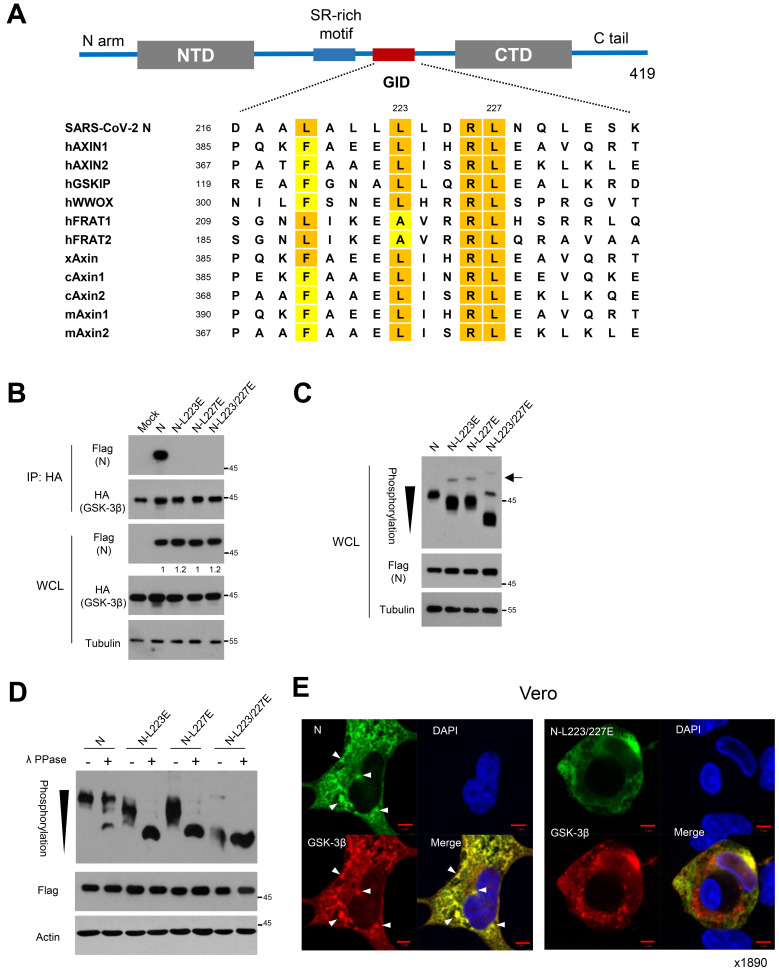
Fig. 1. Potential GID in N of SARS-CoV-2 is responsible for phosphorylation of N.
(A) Schematic diagram of structural organization of SARS-CoV-2 N protein (upper). The Ser/Arg (SR)-rich motif and potential GID are located in the central disordered region. Sequence alignment of GID (L/FxxxL/AxxRL) in SARS-CoV-2 N and endogenous GSK-3 interacting proteins with human (h), mouse (m), xenopus (x), and chicken (c). Highlights are shown for conserved hydrophobic residues of Lue, Phe, and Ala (lower). (B) Flag-tagged N and mutants harboring Lue to Glu substitutions with HA-tagged GSK-3β were expressed in 293 cells. Interaction between N or mutants and GSK-3β were determined following immunoprecipitation (IP) with anti-HA beads. Whole cell lysate (WCL) serve as input abundance for IP. (C) Ancestral or mutant N expression vectors were transfected into 293 cells, and the lysates were subjected to western blot and Phos-tag gel analysis to determine protein abundance and mobility shift by phosphorylation status of N, respectively. Black arrowhead indicates number of phosphorylation residues of N on a Phos-tag gel. Black arrow indicates minor bands of hyperphosphoryation of N mutants in Phos-tag analysis. (D) N or mutant N lysates were subjected to lambda protein phosphatase (λ PPase) followed by Phos-tag gel analysis. Black arrowhead indicates number of phosphorylation residues of N on a Phos-tag analysis. (E) Confocal images of ancestral N or L223/227E mutant (green) and GSK-3β (red) in Vero cells. Arrowheads indicate co-localized foci of N and GSK-3β in condensate-like structures. Nuclear staining with DAPI (blue). Scale bars = 5 μm.
MATERIALS AND METHODS
Expression constructs and antibodies
The 293 cells obtained from ATCC (USA) and Vero cells from Korean Cell Line Bank (Korea) were routinely cultured in DMEM and RPMI1640 medium containing 10% fetal bovine serum, respectively. The expression vector pGBW-m4134490 (plasmid No. 152580) having codon optimized (due to quarantine concern rather than protein expression) N of SARS-CoV-2, pGBW-m4134909 (plasmid No. 151901) having N of human coronavirus 229E, pGBW-m4134899 (plasmid No. 151902) having N of human coronavirus OC43 and pGBW-m4134901 (plasmid No. 151922) having N of human coronavirus HKU1 229E, HA-tagged spike (S) of SARS-CoV-2 (plasmid No. 152113), HA-tagged envelop (E) of SARS-CoV-2 (plasmid No. 153661), and HA-tagged membrane (M) of SARS-CoV-2 (plasmid No. 152583) were obtained from Addgene (USA). Those N expression vectors were subcloned into pcDNA3.1 with C-terminal flag or EGFP tag. Mutant expression vectors of N in GID, linker, and CDK1 phosphorylation site were generated by a polymerase chain reaction-based method. The transfection was performed by Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen, USA). Antibodies against GSK-3β (610202; BD Transduction Laboratories, USA), pS9-GSK-3β (9323S; Cell Signaling Technology, USA), pY216-GSK-3β (612312; BD Transduction Laboratories), Flag (F-3165; Sigma, USA), Snail (L70G2; Cell Signaling Technology), β-catenin (610154; BD Transduction Laboratories), and Tubulin (LF-PA0146; AbFrontier, Korea) were obtained from the commercial vendors. Endogenous GSK-3β kinase activity was determined by commercial kit (V1991; Promega, USA).
Western blot and Phos-tag gel analysis
For the western blot analyses, protein extracts were prepared in Triton X-100 lysis buffer. Phosphorylation status of N protein was determined by anti-flag antibody and mobility shift on a Phos-tag gel (Wako, Japan) as described previously (Lee et al., 2018).
Immunoprecipitation and immunofluorescence
For immunoprecipitation analysis, whole cell Triton X-100 lysates were incubated with Flag-M2 agarose (Sigma) and washed with lysis buffer three times. The recovered proteins were resolved by SDS-PAGE and subjected to immunoblot analysis. For immunofluorescence study, the cells were washed twice with ice-cold phosphate-buffered saline (PBS) and incubated for 15 min at room temperature with 3% formaldehyde in PBS. The cells were permeabilized with 0.5% Triton X-100 for 5 min and then blocked for 1 h in PBS containing 3% bovine serum albumin followed by incubation with primary antibody overnight at 4°C. Cells were then washed three times with PBS containing 0.1% Tween 20 followed by incubation with anti-mouse-Alexa Fluor-488 (for green) or anti-rabbit-Alexa Fluor-594 (for red) secondary antibody. Cellular fluorescence was monitored using confocal microscopy (Zeiss, Germany).
TCF/LEF (T-cell factor/lymphoid enhancer factor) and E-cadherin reporter assay
For TCF/LEF and E-cadherin reporter assay, the cells were transfected with 50 ng of the Super-Top or E-cad(-108)-Luc reporter vector (Kim et al., 2011; Yook et al., 2005) and 1 ng of pSV40-Renilla expression vector in combination with N or N-mutant as indicated. Luciferase and renilla activities were measured using the dual-luciferase reporter system kit (Promega) and the luciferase activity was normalized with renilla activity. The results are expressed as the averages of the ratios of the reporter activities from triplicate experiments.
Split superfolder green fluorescent protein (GFP) assay
For split superfolder GFP assay, the GFP1-10 and GFP11 constructs were kindly provided by Professor Hye Yoon Park at Seoul National University. The GFP1-10 fragment was fused into the N-terminus of GSK-3 and GFP11 was fused into the C-terminus of N. The 293 cells were co-transfected with split GFP vectors, and fluorescence intensity was determined by 5 random areas with the same exposure, followed by ImageJ analysis.
VLP experiment and transmission electron microscopy (TEM)
For the VLP experiment, 293 cells were transfected with HA-tagged E, M and S expression vectors with empty or wild type (wt) N or N-L223/227E expression vectors. At 48 h, the culture supernatant was harvested and cleared by centrifuge (800 rpm for 3 min) followed by 0.2 μm syringe filtration. The supernatant was incubated with Lenti-X concentrator (Clontech Laboratories, Japan) for 16 h followed by centrifugation (15,000 g for 45 min) according to the manufacturer’s instructions. The re-suspended viral pellets in PBS were subjected to western blot and TEM analysis. For negative stain, purified VLPs were placed on Formva-carbon coated grids and the grids were stained with 1% uranyl acetate for 15 s followed by washing with distilled water. Dried grids were observed with TEMT (JEM-1011; JEOL, Japan) at an acceleration voltage of 80 kV and images with particle size were obtained from a TEM-equipped Megaview III CCD camera.
Protein sequences of coronaviruses and SARS-CoV-2 variants
Amino acid sequences of N for SARS-CoV-2 (P0DTC9), SARS-CoV (P59595), HKU1 (Q0ZME3), HCoV-OC43 (P33469), HCoV-229E (PP15130), HCoV-NL63 (Q6Q1R8) were obtained from UniProt (https://www.uniprot.org/). Mutational information on N protein in SARS-CoV-2 variants was obtained from PANGO Lineages (https://cov-lineages.org/constellations).
Statistics
Statistical analysis for reporter, GSK-3 kinase, and split GFP assay was performed with two-tailed Student’s t-test; data are expressed as means and SD. IBM SPSS Statistics (released 2017 for Windows, ver. 25.0; IBM, USA) was used for all statistical analyses. The double asterisks denote P < 0.01, one asterisk denoting P < 0.05.
RESULTS
N of SARS-CoV-2 harbors a GSK-3 interacting domain (GID) similar to that of AXIN
Phosphorylation of many endogenous and viral proteins by GSK-3 is dependent on GSK-3 binding of protein-protein interaction (Dajani et al., 2003; Fujimuro et al., 2005). In turn, the GID embedded in the GSK-3 substrate provides efficient self-phosphorylation. Because N of coronavirus is phosphorylated by GSK-3 (Peng et al., 2008; Wu et al., 2009), we examined whether GID provides N-GSK-3 interaction and efficient phosphorylation, finding a L/FxxxLxxRL motif in the unstructured disordered region of ancestral Wuhan Hu-1 N protein (Fig. 1A). The amino acid sequence of GID is highly conserved in many GSK-3 interacting proteins, such as AXINs, GSKIP and WWOX. Because the hydrophobic residues in GID play a critical role in GSK-3 interaction (Ahn et al., 2017a; 2017b; Dajani et al., 2003), we generated Leu to Glu substitution mutants in GID (L223/227E) from ancestral SARS-CoV-2 N protein. We next transfected them with HA-tagged GSK-3β and N expression vectors in 293 cells and performed immunoprecipitation assay to examine interaction between N and GSK-3. Indeed, N protein interacts with GSK-3 and point mutant of hydrophobic residues in N-GID, largely abolishing the interaction with GSK-3, although protein abundance of N and mutants was comparable (Fig. 1B). To further determine the role of N-GSK-3 interaction, we next examined the phosphorylation status of N and GID mutants by means of mobility shift on a Phos-tag gel (phosphate-affinity SDS-PAGE). The Zn2+ in Phos-tag gel decreases mobility shift of phosphorylated residues, providing a tool to examine phosphorylation status without phospho-specific antibody (Kinoshita et al., 2006). In Phos-tag analysis, mutation of hydrophobic residue(s) largely decreased phosphorylation of N (i.e., increased mobility shifts) without affecting its stability (Fig. 1C). The N mutants, but not of wt N, revealed a minor band of hyperphosphoryation in Phos-tag analysis, suggesting that other endogenous kinases may be involved in phosphorylation of N mutants. The GID mutation in Axin2 scaffolding protein largely abolished Axin2 phosphorylation along with protein abundance (Supplementary Fig. S1A) (Yamamoto et al., 1999). To further determine the phosphorylation status of N and its mutants, we treated it with lambda protein phosphatase (λ PPase) and subjected it to Phos-tag gel. The λ PPase treatment increased mobility shift of N and point mutant of L223E or L227E, while the L223/227E mutation was unaffected (Fig. 1D). Interestingly, mobility shift of N-L223/227E mutant was not affected by λ PPase treatment, suggesting that GID provides the main N phosphorylation. Immunofluorescence also supported the co-localization of N and GSK-3, especially in condensate-like structures in the cytosolic space, as compared to L223/227E mutant N in Vero as well as 293 cells (Fig. 1E, Supplementary Fig. S1B). Our observations indicate that N in SARS-CoV-2 harbors GID, providing interaction with GSK-3 followed by enhanced phosphorylation of N by GSK-3.
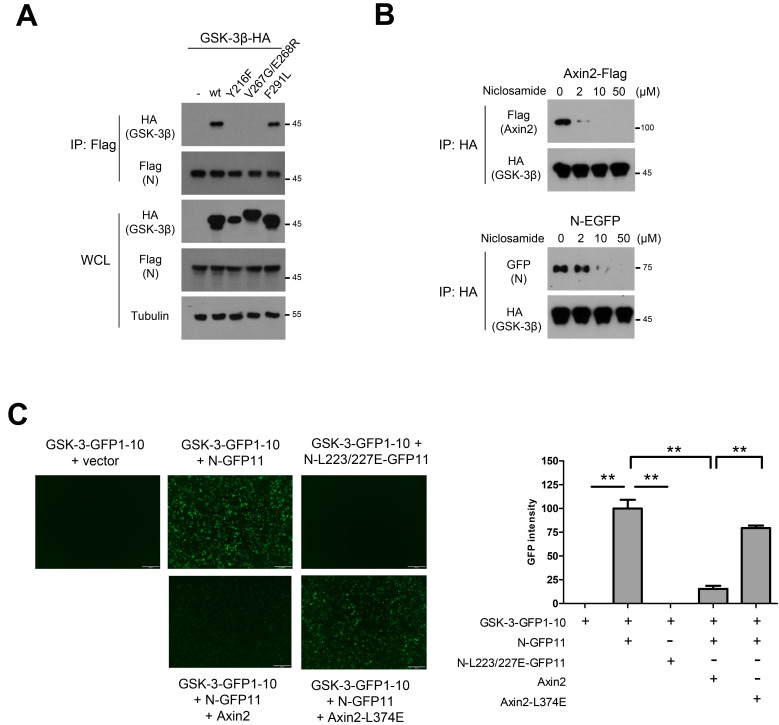
Because the Axin-GSK-3 binding structure is well-determined (Dajani et al., 2003; Fraser et al., 2002), we next compared the binding site of Axin2 and N with that in wt and mutants GSK-3. Consistently, Y216 and V267/E268 in GSK-3 were critical for Axin2 binding (Supplementary Fig. S2), and N shared those binding sites in GSK-3 (Fig. 2A), indicating that N utilizes a GSK-3 binding mode similar to that of Axin. Previously, we reported that antihelminthic niclosamide interacts with GSK-3 and inhibits Axin-GSK3 binding, resulting in attenuation of Wnt signaling and EMT in APC-mutated colorectal cancer (Ahn et al., 2017a). Niclosamide also inhibits viral replication of SARS-CoV-1 in Vero E6 cells (Wu et al., 2004) although the molecular mechanism of action (MoA) of niclosamide on SARS-CoV is unclear. Given the similar binding modes of N and Axin to GSK-3, we next performed an in vitro immunoprecipitation assay with increasing doses of niclosamide into the cell lysate. Indeed, niclosamide inhibited Axin2 or N binding to GSK-3 in a dose-dependent manner (Fig. 2B). While N-GSK-3 interaction was maintained at 2 μM concentration of niclosamide, Axin2 binding to GSK-3 was largely abolished at the same concentration. These results suggest that N harbors stronger binding affinity to GSK-3 than does Axin2 and explain the modest therapeutic potential of niclosamide in SARS-CoV-2 in vitro (Ko et al., 2021). To test whether N and Axin share the GSK-3 binding site, we next utilized split GFP systems (Cabantous et al., 2005). For the split GFP assay, superfolder GFP1-10 (amino acids 1-214) and GFP11 (amino acid 214-230) fragments were fused to GSK-3 and N, respectively. Co-transfection of GSK3-GFP1-10 and N-GFP11, but not L223/227E mutant, gave rise to green fluorescence via direct interaction between GSK-3 and N. The fluorescence was largely decreased by the overexpression of Axin2 while it was slightly decreased by the overexpression of Axin2-L374E mutant, indicating that N and Axin compete in similar binding regions of GSK-3 (Fig. 2C).
Fig. 2. N interacts with GSK-3β similarly to Axin2.
(A) Flag-tagged N was co-transfected with wt (wild type) or Axin-binding mutant (Y216F, V267G/E268R, F291L) GSK-3β expression vectors in 293 cells. Following immunoprecipitation (IP) with anti-flag, GSK-3β binding was determined by western blot analysis. WCL, whole cell lysate. (B) Flag-tagged N was co-transfected with HA-tagged wt GSK-3β in 293 cells. The lysates were subjected to IP with increasing doses of niclosamide in vitro, and dissociation of Axin2 or N was determined by western blot analysis. (C) One hundred ng of each GSK-3β fused to GFP1-10 and control empty vector (upper left) or N (upper middle) or N-L223/227E mutant (upper right) fused to GFP11 were co-transfected into 293 cells for split GFP analysis. Competition between N and Axin2 was determined by overexpression of 1 μg of each Axin2 (lower left) or Axin2-L374E (lower right). Scale bars = 200 µm. Green fluorescence image was obtained from 5 random fields (×100) and the intensity was quantitatively determined using ImageJ software (right panel). **P < 0.01.
N interaction is irrelevant to GSK-3 activity and canonical Wnt activity
Because Axin directly suppresses kinase activity of GSK-3 via GID, thus regulating various signaling pathways (Cohen and Frame, 2001; Doble and Woodgett, 2003), we next examined the role of N in GSK-3 kinase activity and the subsequent Wnt/EMT signaling pathway. Surprisingly, overexpression of N or N-GID mutant did not affect endogenous GSK-3 kinase activity in 293 cells (Supplementary Fig. S3A). Furthermore, overexpression of N or N-GID mutant did not affect GSK-3 abundance or phosphorylation status on Ser9 and Y216 (Supplementary Fig. S3B). Overexpression of N neither increases protein abundance of β-catenin and Snail, nor affects TCF/LEF transcriptional and E-cadherin promoter activity (Supplementary Fig. S3C). These results indicate that N, unlike endogenous GSK-3 scaffolding proteins, utilizes GSK-3 for its phosphorylation but does not affect GSK-3 kinase activity and subsequent signaling pathways.
N phosphorylation is required for structural loading in virus-like particles (VLPs)
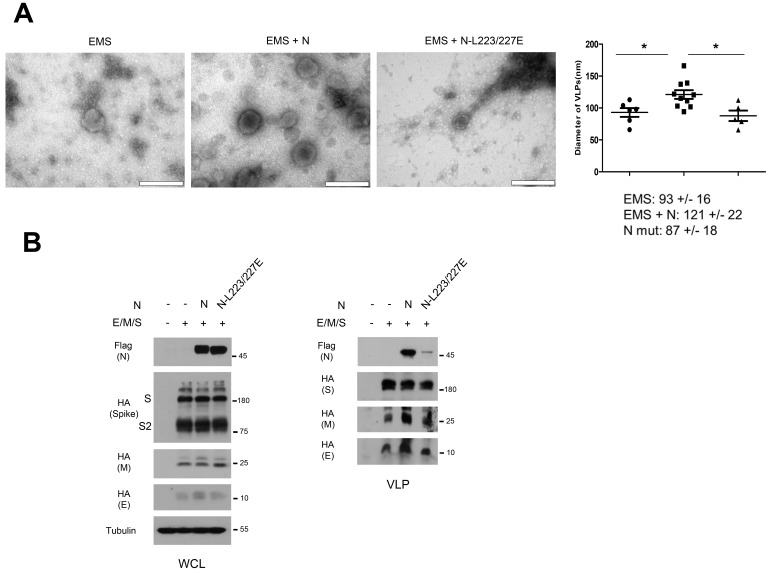
Phosphorylation of N is essentially required for its phase separation and subsequent viral replication (Carlson et al., 2020; Wu et al., 2009; 2014). Because the N comprises an important structural component of a viral particle, we next examined the role of N or N-L223/227E mutant in VLP production. While the VLP can be produced by the envelope (E), membrane (M) and spike (S) without N, electron microscopic analysis revealed that N protein provides a bigger particle size with solid structure of VLP while the N-L223/227E mutant was similar to the absence of N (Fig. 3A). Interestingly, N-L223/227E mutant loading in VLPs was significantly decreased compared to wt N, while protein abundance of N and other structural components were comparable in cell lysate (Fig. 3B). These results indicate that N phosphorylation is also required for structural assembly of viral particles.
Fig. 3. Phosphorylation is required for structural loading of N in virus-like particle (VLP).
(A) The 293 cells were transfected with E, M, S and empty vector (EMS) or EMS with wt N expression vector (EMS + N) or EMS with N-L223/227E expression vector (EMS + N-L223/227E) and VLPs were subjected to TEM analysis (left panels). Scale bars = 200 nm. The VLP sizes of EMS (93 ± 16 nm), EMS + N (121 ± 22 nm) and EMS-N-L223/227E (87 ± 18 nm) were determined by TEM images (right). *P < 0.05 by t-test. (B) Cell lysate (left) for VLP production in 293 cells and purified VLP (right) were subjected to western blot analysis to determine HA-tagged E, M, S, and flag-tagged N or N-L223/227E. S2, S2 subunit of S; WCL, whole cell lysate.
N-GID evolution of Sarbecovirus lineage of coronavirus
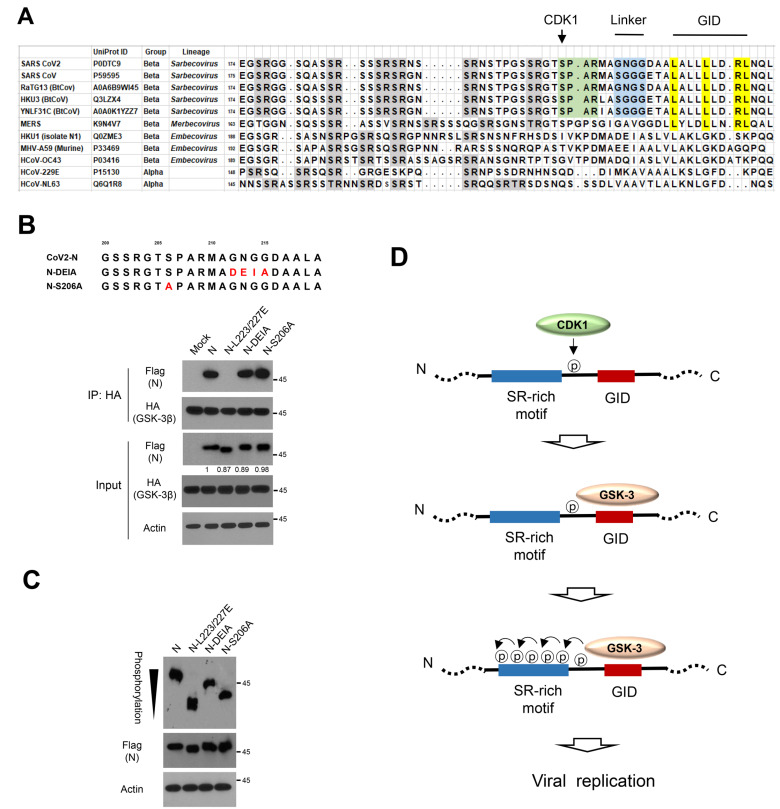
While human coronaviruses HKU1, OC43, 229E, and NL63 typically cause upper respiratory infection with relatively minor symptoms, SARS-CoV and SARS-CoV-2 in the Sarbecovirus lineage of beta-coronavirus replicate in the lower respiratory tract and cause severe pneumonia (Tay et al., 2020). To understand the role of GID in the evolution of coronavirus, we next analyzed GID and SR-rich phosphorylation motif among various coronaviruses, including bat coronavirus of RaTG13, which is associated with the SARS-CoV-2 (Zhou et al., 2020) (Fig. 4A). Intriguingly, we found several differences in N-GID of Sarbecovirus lineage SARS-CoV-2 as compared to other coronaviruses (Tabibzadeh et al., 2021). First, Sarbecovirus lineage N including RaTG13 harbors a typical GID, similar to those of endogenous GSK-3 binding proteins. Second, N of Sarbecovirus contained a CDK1 phosphorylation site (S206) with a conserved pSPxK/R motif that may serve to prime phosphorylation of GSK-3 (Surjit et al., 2006). Third, N of Sarbecovirus has a Gly-rich flexible linker between GID and SR-rich phosphorylation domain. It should be noted that MERS-CoV in Merbecovirus lineage has a GID and Gly-rich linker similarly to Sarbecovirus, but it lacks typical CDK-1 phosphorylation for GSK-3. Based on these observations, we hypothesize that CDK1 serves to prime phosphorylation of GSK-3 and that Gly-rich linker facilitates phosphorylation of N. To test whether the CDK1 phosphorylation site and Gly-rich linker enhance phosphorylation of N by GSK-3, we generated N expression vectors with substitution either of CDK1 phosphorylation site (S206A) or GNGG linker to DEIA (N-DEIA), corresponding to sequences in the HKU1 coronavirus (Fig. 4B). Then, we transfected the expression vectors and examined the protein abundance, GSK-3 binding and phosphorylation status of N. We found a similar protein abundance and binding capacity to GSK-3 of N and mutant N expression vectors while the GID mutant (N-L223/227E) was unbound to GSK-3 (Fig. 4B). Interestingly, mutation of the CDK1 phosphorylation site (N-S206A) and of Gly-rich linker (N-DEIA) decreased phosphorylation status migrating between ancestral N and GID mutant (N-L223/227E mutant) in Phos-tag analysis (Fig. 4C). These results indicate that emergence of CDK1 and Gly-rich linker together with GID in the bat Sarbecovirus lineage play an important role in enhanced phosphorylation of N by GSK-3 (Fig. 4D).
Fig. 4. CDK1-primed phosphorylation and Gly-rich linker enhance N phosphorylation.
(A) The N sequence of the Ser/Arg (SR)-rich motif and GSK-3 interacting domain (GID) in various coronavirus groups including bat RaTG13 causing human respiratory infection. Green indicates CDK1 phosphorylation site, blue indicates Gly-rich linker, gray indicates SR-rich motif and yellow indicates GID. (B) Sequence alignment of N expression vectors having CDK1 phosphorylation site mutant (S206A) and substitution of Gly-rich linker (from aa 212-215) to DEIA found in HKU1 coronavirus (upper). The ancestral N or N-DEIA or N-S206A were co-transfected with HA-tagged GSK-3β into 293 cells, and the lysates were subjected to immunoprecipitation (IP) with anti-HA and subsequent western blot analysis to determine N binding affinity. (C) The ancestral N or N-DEIA or N-S206A were transfected into 293 cells and the lysates were subjected to Phos-tag gel analysis to determine N phosphorylation status. Black arrowhead indicates number of phosphorylation residues of N on a Phos-tag analysis. (D) Schematic diagram of CDK1 priming phosphorylation followed by GSK-3-mediated serial phosphorylation of N.
Phosphorylation of N in other coronavirus and SARS-CoV-2 variants
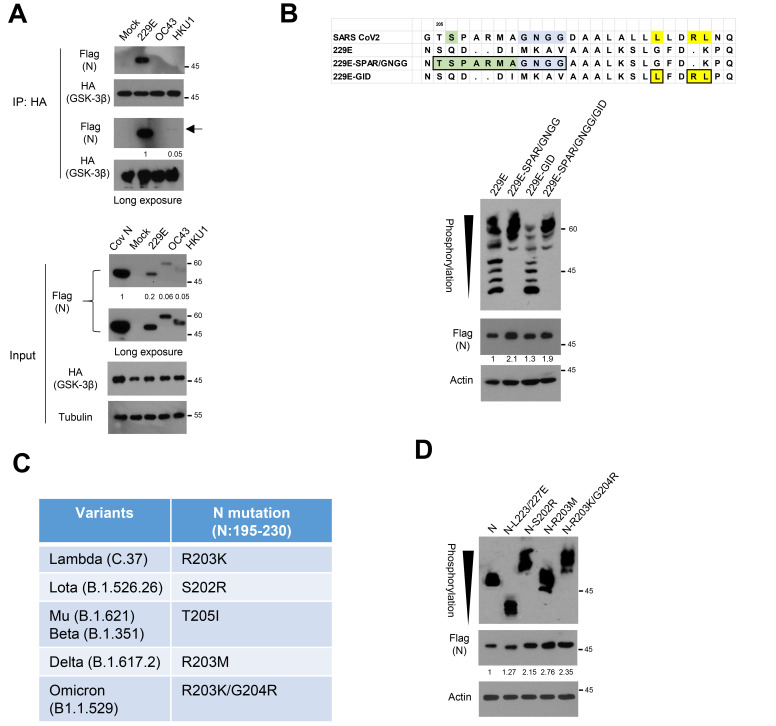
Given the difference between the Sarbecovirus lineage of beta-coronavirus and other coronaviruses, we next compared N abundance in those coronaviruses. Transfecting N of SARS-CoV-2, 229E, OC43, and HKU1 strains and examining protein abundance by western blot analysis, we found significantly increased N abundance in SARS-CoV-2 compared to N in other coronaviruses (Fig. 5A). Interestingly, 229E-N revealed detectable binding to GSK-3 while N-HKU1 or N-OC43 showed much weaker or absence of interaction, respectively. To further determine the role of GSK-3 and GID, we next generated 229E-N expression vectors having a CDK1-primed phosphorylation site, Gly-rich linker, and GID corresponding to those of sequences in N of SARS-CoV-2. We found that introduction of primed phosphorylation and Gly-rich linker significantly increased phosphorylation as well as protein abundance (about 2-fold) compared to wt N-229E (Fig. 5B). In Phos-tag analysis, multiple phosphorylation states can be determined by the laddering effect because each phosphorylated Ser/Thr protein appears in a different band (Kinoshita et al., 2006). Notably, wt 229E-N showed at least 7 bands in Phos-tag gel analysis, the less-phosphorylated bands disappearing with the introduction of a primed phosphorylation site and flexible linker between the SR-rich motif and GID. Because the GSK-3 phosphorylates N up to 10 phosphates in the SR-rich motif (Carlson et al., 2020; Wu et al., 2009; 2014), our observation indicates that SR-rich motif in wt 229E-N is not fully phosphorylated by GSK-3. Although 229E-N does not have a typical GID of N-SARS-CoV-2, substitution of hydrophobic residues largely abolished GSK-3 interaction and phosphorylation status (Supplementary Fig. S4). These results support that the GID, CDK-1 mediated primed phosphorylation, and flexible linker between GID and the SR-rich motif contribute to the N hyper-phosphorylation of coronavirus.
Fig. 5. N in other coronaviruses and N variants.
(A) The ancestral N or N from 229E, OC43, and HKU1 were co-transfected with HA-tagged GSK-3β into 293 cells, and the lysates were subjected to immunoprecipitation (IP) with anti-HA and subsequent western blot analysis to determine N binding affinity. Relative binding affinity of 229E and HKU1 corresponding each input was measured in a long exposure film. (B) Sequence alignment of 229E-N expression vectors having CDK1 phosphorylation site and Gly-rich linker (SPAR/GNGG) or GSK-3 interacting domain (GID) found in SARS-CoV-2 N (upper). The 229E-N or mutants were transfected into 293 cells. The lysates were subjected to western blot and Phos-tag gel analysis to determine protein abundance and mobility shift by phosphorylation status of 229E-N, respectively. Black arrowhead indicates number of phosphorylation residues of N on a Phos-tag analysis. (C) Mutation profiles of N between SR-rich motif and GID (N195-230) in SARS-CoV-2 variants (https://cov-lineages.org/constellations). (D) Ancestral N and mutants found in the variants were transfected into 293 cells. The lysates were subjected to western blot and Phos-tag gel analysis to determine protein abundance and phosphorylation status of N and mutants, respectively. Black arrowhead corresponds to the increased phosphorylation in Omicron variant and ancestral N phosphorylation on a Phos-tag gel. N-L223/227E mutant serves as control for un-phosphorylated N.
The continuous evolution of SARS-CoV-2 through mutational changes has given rise to many variants including Delta (B.1.617.2) and Omicron (B.1.1.529). Given our observations, we further examined the N mutation in those variants focusing on the region between the SR-rich motif and GID (https://cov-lineages.org/constellations). Interestingly, we found many mutations in the variants of SARS-CoV-2 in the proximal N-terminus of the CDK1 phosphorylation site (Fig. 5C). Notably, there was no mutation among the SARS-CoV-2 variants in the GID region, suggesting the evolutionary fitness of GSK-3 binding in the Sarbecovirus lineage. To validate the role of these mutations in the variants, we chose several mutations of N including Variants of Concern (VOC, Delta and Omicron) and generated N expression vectors having those mutations. We then overexpressed those vectors in 293 cells and examined the protein abundance and phosphorylation status of N. Interestingly, protein abundance of N in those variants was increased by the substitution of single or double amino acid(s), especially in Delta and Omicron (Fig. 5D). The N-S202R mutant found in Lota (Variant Being Monitored, VOM) and R203K/G204R found in Omicron showed even more phosphorylation compared to ancestral N, while N found in Delta revealed a similar mobility shift. These results indicate that the mutational evolution of N between the SR-rich motif and GID increases protein abundance as well as hyper-phosphorylation of N. Taking these together, we observed that the N protein of SARS-CoV-2 harbors hyper-phosphorylation machinery including GID, priming phosphorylation, and Gly-rich flexible linker. The evolutionary role and biological impact of N in the COVID-19 pandemic thus needs further elucidation.
DISCUSSION
Highly expressed endogenously in mammals, GSK-3 is an exceptional kinase inactivated by exogenous signaling, such as by insulin and Wnt (Cohen and Frame, 2001; Doble and Woodgett, 2003). Priming phosphorylation by other many kinases allows a strong (500 to 1,000 fold) preference for GSK-3 substrates, which have multiple and serial phosphorylation sites by GSK-3, including EMT-inducer Snail and β-catenin (Kaidanovich-Beilin and Woodgett, 2011; Yook et al., 2005). The importance of GSK-3 has been extensively confirmed in human diseases, including Alzheimer’s, metabolic diseases, and cancer (Cohen and Frame, 2001; Doble and Woodgett, 2003). Notably, many viruses or bacterial genes directly interact with and utilize endogenous GSK-3 for entry, replication and latency (Alfhili et al., 2020). For example, latency-associated nuclear antigen (LANA) in Kaposi’s sarcoma-associated herpesvirus (KSHV) interacts with GSK-3 via GID embedded in LANA. As with Axin, the LANA-GSK-3 interaction is essential to phosphorylation of LANA and inactivation of GSK-3 (Fujimuro et al., 2005). Cytotoxin-associated gene A (CagA) oncoprotein in Helicobacter pylori directly binds to GSK-3 and depletes its endogenous kinase activity, resulting in potentiation of the Snail-mediated EMT program in gastric cancer (Lee et al., 2014). Unlike LANA and CagA, N of SARS-CoV-2 does not affect endogenous GSK-3 activity. Because the endogenous GSK-3 kinase activity is critically required for replication of coronavirus, including phase separation, RNA transcription and viral particle formation (Carlson et al., 2020; Wu et al., 2009; 2014), our observations indicate that N-GID has evolved to facilitate its own phosphorylation as a GSK-3 substrate rather than as a GSK-3 scaffolding protein in Wnt signaling.
SARS-CoV-2 is classified as beta-coronaviruses genus and Sarbecovirus lineage (lineage B), with a close phylogenetic relationship to bat coronavirus RaTG13 (Zhou et al., 2020). The multifunctional N protein of the coronavirus family not only constitutes a key structural protein of virion but also plays a critical role in RNA binding, packaging, and other replication processes (Chang et al., 2014). Because the N protein is abundantly produced during the early phases of viral replication of SARS-CoV-2 as well as SARS-CoV-1 (Meyer et al., 2014; Shan et al., 2021; Tan et al., 2004), our observation suggests that N protein abundance and subsequent post-translation modification play an important role in early stage of human infection. Based on previous observations that GSK-3 phosphorylation of N is critically important for viral RNA binding (Chen et al., 2005; Lu et al., 2021; Wu et al., 2009), we queried the role of GSK-3-mediated phosphorylation of N during SARS-CoV-2 evolution because GSK-3 comprises a key kinase involving many endogenous biological events (Cohen and Frame, 2001; Doble and Woodgett, 2003). We found the GID amino acid sequence in N of Sarbecovirus and Merbecovirus lineage consists of L/FxxxL/AxxRL, consistent with the putative alpha helix predicted by Monte Carlo simulation in the flexible disordered linker region (Cubuk et al., 2021). Axin binds to GSK-3 via alpha helical GID, followed by formation of the APC and β-catenin complex, which is critically important for phosphorylation and protein abundance of Axin (Yamamoto et al., 1999). The substitution of hydrophobic residues in N-GID (L223/227E), which plays a critical role in GSK-3 binding, largely abolished the interaction with GSK-3 without affecting N protein abundance. Our observations suggest that N, unlike GSK-3 scaffolding proteins, is highly phosphorylated by GSK-3. Since the phosphorylation of N is also critically important for self-oligomerization, multivalent RNA-protein complex formation, and subsequent liquid-liquid separation with RNA condensates formation (Carlson et al., 2020; Lu et al., 2021), our observations suggest that N-GID may provide replication advantage of coronavirus. We also found that N phosphorylation by GSK-3 is required for structural N in viral particles as determined by VLP experiment. The phosphorylation status of N protein in SARS-CoV-2 virion, role of GID-mediated N phosphorylation in viral particle formation, and existence of endogenous protein phosphatase are thus interesting issues raised by this study.
While SARS-CoV-2 is currently evolving increased transmissibility, the functional importance of mutations in the variants is technically challenging. Recent observation has revealed that S202R in Lota and R203M in Delta variant provide 166-fold and 51-fold higher viral production, respectively (Syed et al., 2021). In this study, we showed that pS206 priming phosphorylation by CDK1 and Gly-rich linker located between GID and SR-rich motif enhance phosphorylation of N. Given the multiple bands in N of the 229E strain and high density of up to 10 phosphates in the SR-rich motif in various strains of coronavirus, N of SARS-CoV-2 also has multiple phosphorylation sites in the SR-rich motif. Interestingly, emerging variants have mutations near the primed phosphorylation site. Notably, R203K/G204R, found in a highly transmissible Omicron variant, allows greater phosphorylation and protein abundance than N of ancestral and other variants. Because more than fourteen Ser/Thr residues exist in the SR-rich motif in Sarbecovirus lineage, our results indicate that N of Omicron gains further phosphorylation via R203K/G204R mutation. Taken together, our observations suggest that enhanced phosphorylation and increased protein abundance of N as well as mutations of Spike may provide a selective advantage during the mutational evolution of SARS-CoV-2. Further study is needed to elucidate the importance of N phosphorylation in the emergence and evolution of SARS-CoV-2.
Previously, we observed that the anthelmintic niclosamide disrupts Axin-GSK3 interaction, providing a repositioned therapeutic for colon cancer and familial adenomatosis coli (Ahn et al., 2017b). While niclosamide is consistently effective in disrupting Axin2-GSK-3 binding, an at least 5-fold concentration of niclosamide was required for disruption of N-GSK-3 interaction in our hands, indicating that N has a stronger interaction with GSK-3 than does Axin2. Conversely, our observations provide an MoA of niclosamide on SARS-CoV replication and N expression, at least in part (Wu et al., 2004), with implications for clinical trials of niclosamide for SARS-CoV-2 (Al-Kuraishy et al., 2021). Although GSK-3 kinase inhibitors can be a therapeutic target for SARS-CoV-2, the endogenous abundance of GSK-3 along with its diverse physiological roles largely limit the therapeutic potential of GSK-3 inhibitors for viral diseases. Thus, further study is required regarding the protein-protein interaction of N-GSK-3 as a therapeutic target for SARS-CoV-2 infection in human.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
We thank E. Tunkle for preparation of the manuscript and J. Choi at Dongduk University College of Pharmacy for technical assistance. This work was supported by grants from the National Research Foundation of Korea (NRF-2019R1A2C2084535, NRF-2021R1A2C3003496, NRF-2022R1A2C3004609) funded by the Korean government (MSIP), and a grant from the National Research Foundation of Korea (NRF-2020R1I1A1A01072977) funded by the Korean government (MOE).
Footnotes
AUTHOR CONTRIBUTIONS
J.S.Y. and H.S. performed all experiments. S.Y.C., J.E.L., C.-H.J., S.H.S., S.K., and E.S.C. supported experiments. N.H.K. and K.H.H. performed the split GFP assay. N.H.K., H.S.K., and J.I.Y. planned all experiments, analyzed the data, and wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ahn S.Y., Kim N.H., Lee K., Cha Y.H., Yang J.H., Cha S.Y., Cho E.S., Lee Y., Cha J.S., Cho H.S., et al. Niclosamide is a potential therapeutic for familial adenomatosis polyposis by disrupting Axin-GSK3 interaction. Oncotarget. 2017a;8:31842–31855. doi: 10.18632/oncotarget.16252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S.Y., Yang J.H., Kim N.H., Lee K., Cha Y.H., Yun J.S., Kang H.E., Lee Y., Choi J., Kim H.S., et al. Anti-helminthic niclosamide inhibits Ras-driven oncogenic transformation via activation of GSK-3. Oncotarget. 2017b;8:31856–31863. doi: 10.18632/oncotarget.16255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfhili M.A., Alsughayyir J., McCubrey J.A., Akula S.M. GSK-3-associated signaling is crucial to virus infection of cells. Biochim. Biophys. Acta Mol. Cell Res. 2020;1867:118767. doi: 10.1016/j.bbamcr.2020.118767. [DOI] [PubMed] [Google Scholar]
- Al-Kuraishy H.M., Al-Gareeb A.I., Alzahrani K.J., Alexiou A., Batiha G.E. Niclosamide for Covid-19: bridging the gap. Mol. Biol. Rep. 2021;48:8195–8202. doi: 10.1007/s11033-021-06770-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabantous S., Terwilliger T.C., Waldo G.S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- Carlson C.R., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Safari M., Frankel A.D., Morgan D.O. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. Mol. Cell. 2020;80:1092–1103.e4. doi: 10.1016/j.molcel.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein--forms and functions. Antiviral Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Gill A., Dove B.K., Emmett S.R., Kemp C.F., Ritchie M.A., Dee M., Hiscox J.A. Mass spectroscopic characterization of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding by using surface plasmon resonance. J. Virol. 2005;79:1164–1179. doi: 10.1128/JVI.79.2.1164-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P., Frame S. The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021;12:1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajani R., Fraser E., Roe S.M., Yeo M., Good V.M., Thompson V., Dale T.C., Pearl L.H. Structural basis for recruitment of glycogen synthase kinase 3beta to the axin-APC scaffold complex. EMBO J. 2003;22:494–501. doi: 10.1093/emboj/cdg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Rottier P.J. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doble B.W., Woodgett J.R. GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser E., Young N., Dajani R., Franca-Koh J., Ryves J., Williams R.S., Yeo M., Webster M.T., Richardson C., Smalley M.J., et al. Identification of the Axin and Frat binding region of glycogen synthase kinase-3. J. Biol. Chem. 2002;277:2176–2185. doi: 10.1074/jbc.M109462200. [DOI] [PubMed] [Google Scholar]
- Fujimuro M., Liu J., Zhu J., Yokosawa H., Hayward S.D. Regulation of the interaction between glycogen synthase kinase 3 and the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen. J. Virol. 2005;79:10429–10441. doi: 10.1128/JVI.79.16.10429-10441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgepeth C.M., Deardorff M.A., Rankin K., Klein P.S. Regulation of glycogen synthase kinase 3beta and downstream Wnt signaling by axin. Mol. Cell. Biol. 1999;19:7147–7157. doi: 10.1128/MCB.19.10.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howng S.L., Hwang C.C., Hsu C.Y., Hsu M.Y., Teng C.Y., Chou C.H., Lee M.F., Wu C.H., Chiou S.J., Lieu A.S., et al. Involvement of the residues of GSKIP, AxinGID, and FRATtide in their binding with GSK3beta to unravel a novel C-terminal scaffold-binding region. Mol. Cell. Biochem. 2010;339:23–33. doi: 10.1007/s11010-009-0366-0. [DOI] [PubMed] [Google Scholar]
- Kaidanovich-Beilin O., Woodgett J.R. GSK-3: functional insights from cell biology and animal models. Front. Mol. Neurosci. 2011;4:40. doi: 10.3389/fnmol.2011.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.H., Kim H.S., Li X.Y., Lee I., Choi H.S., Kang S.E., Cha S.Y., Ryu J.K., Yoon D., Fearon E.R., et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J. Cell Biol. 2011;195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Ko M., Jeon S., Ryu W.S., Kim S. Comparative analysis of antiviral efficacy of FDA-approved drugs against SARS-CoV-2 in human lung cells. J. Med. Virol. 2021;93:1403–1408. doi: 10.1002/jmv.26397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutikov M., Palmer T., Tut G., Fuller C., Shrotri M., Williams H., Davies D., Irwin-Singer A., Robson J., Hayward A., et al. Incidence of SARS-CoV-2 infection according to baseline antibody status in staff and residents of 100 long-term care facilities (VIVALDI): a prospective cohort study. Lancet Healthy Longev. 2021;2:e362–e370. doi: 10.1016/S2666-7568(21)00093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.G., Kim H.S., Lee Y.S., Kim S., Cha S.Y., Ota I., Kim N.H., Cha Y.H., Yang D.H., Lee Y., et al. Helicobacter pylori CagA promotes Snail-mediated epithelial-mesenchymal transition by reducing GSK-3 activity. Nat. Commun. 2014;5:4423. doi: 10.1038/ncomms5423. [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim N.H., Cho E.S., Yang J.H., Cha Y.H., Kang H.E., Yun J.S., Cho S.B., Lee S.H., Paclikova P., et al. Dishevelled has a YAP nuclear export function in a tumor suppressor context-dependent manner. Nat. Commun. 2018;9:2301. doi: 10.1038/s41467-018-04757-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Verma A., Garcia G., Jr., Ramage H., Jr., Lucas A., Jr., Myers R.L., Jr., Michaelson J.J., Jr., Coryell W., Jr., Kumar A., Jr., Charney A.W., Jr., et al. Targeting the coronavirus nucleocapsid protein through GSK-3 inhibition. Proc. Natl. Acad. Sci. U. S. A. 2021;118:e2113401118. doi: 10.1073/pnas.2113401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Ye Q., Singh D., Cao Y., Diedrich J.K., Yates J.R., 3rd, Villa E., 3rd, Cleveland D.W., 3rd, Corbett K.D., 3rd The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun. 2021;12:502. doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T.Y., Lee K.R., Tarn W.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008;275:4152–4163. doi: 10.1111/j.1742-4658.2008.06564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Woo H.G. Molecular perspectives of SARS-CoV-2: pathology, immune evasion, and therapeutic interventions. Mol. Cells. 2021;44:408–421. doi: 10.14348/molcells.2021.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D., Johnson J.M., Fernandes S.C., Suib H., Hwang S., Wuelfing D., Mendes M., Holdridge M., Burke E.M., Beauregard K., et al. N-protein presents early in blood, dried blood and saliva during asymptomatic and symptomatic SARS-CoV-2 infection. Nat. Commun. 2021;12:1931. doi: 10.1038/s41467-021-22072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Chow V.T., Lal S.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed A.M., Taha T.Y., Tabata T., Chen I.P., Ciling A., Khalid M.M., Sreekumar B., Chen P.Y., Hayashi J.M., Soczek K.M., et al. Rapid assessment of SARS-CoV-2-evolved variants using virus-like particles. Science. 2021;374:1626–1632. doi: 10.1126/science.abl6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabibzadeh A., Esghaei M., Soltani S., Yousefi P., Taherizadeh M., Safarnezhad Tameshkel F., Golahdooz M., Panahi M., Ajdarkosh H., Zamani F., et al. Evolutionary study of COVID-19, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as an emerging coronavirus: phylogenetic analysis and literature review. Vet. Med. Sci. 2021;7:559–571. doi: 10.1002/vms3.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Goh P.Y., Fielding B.C., Shen S., Chou C.F., Fu J.L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E., et al. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Y., Juo L.I., Lin Y.T., Hsiao M., Lin J.T., Tsai C.H., Tzeng Y.H., Chuang Y.C., Chang N.S., Yang C.N., et al. WW domain-containing oxidoreductase promotes neuronal differentiation via negative regulation of glycogen synthase kinase 3β. Cell Death Differ. 2012;19:1049–1059. doi: 10.1038/cdd.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Chen P.J., Yeh S.H. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe. 2014;16:462–472. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Yeh S.H., Tsay Y.G., Shieh Y.H., Kao C.L., Chen Y.S., Wang S.H., Kuo T.J., Chen D.S., Chen P.J. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J. Biol. Chem. 2009;284:5229–5239. doi: 10.1074/jbc.M805747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.J., Jan J.T., Chen C.M., Hsieh H.P., Hwang D.R., Liu H.W., Liu C.Y., Huang H.W., Chen S.C., Hong C.F., et al. Inhibition of severe acute respiratory syndrome coronavirus replication by niclosamide. Antimicrob. Agents Chemother. 2004;48:2693–2696. doi: 10.1128/AAC.48.7.2693-2696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Kishida S., Kishida M., Ikeda S., Takada S., Kikuchi A. Phosphorylation of axin, a Wnt signal negative regulator, by glycogen synthase kinase-3beta regulates its stability. J. Biol. Chem. 1999;274:10681–10684. doi: 10.1074/jbc.274.16.10681. [DOI] [PubMed] [Google Scholar]
- Yook J.I., Li X.Y., Ota I., Fearon E.R., Weiss S.J. Wnt-dependent regulation of the E-cadherin repressor snail. J. Biol. Chem. 2005;280:11740–11748. doi: 10.1074/jbc.M413878200. [DOI] [PubMed] [Google Scholar]
- Yook J.I., Li X.Y., Ota I., Hu C., Kim H.S., Kim N.H., Cha S.Y., Ryu J.K., Choi Y.J., Kim J., et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat. Cell Biol. 2006;8:1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.