Extended Data Fig. 2. Histone modification mark, enhancer reporter assay, and ATAC-seq analysis to examine regulatory DNA element at the GATA3 locus and the effects of rs3824662 genotype in normal human tissues and cells as well as human ALL cell lines.
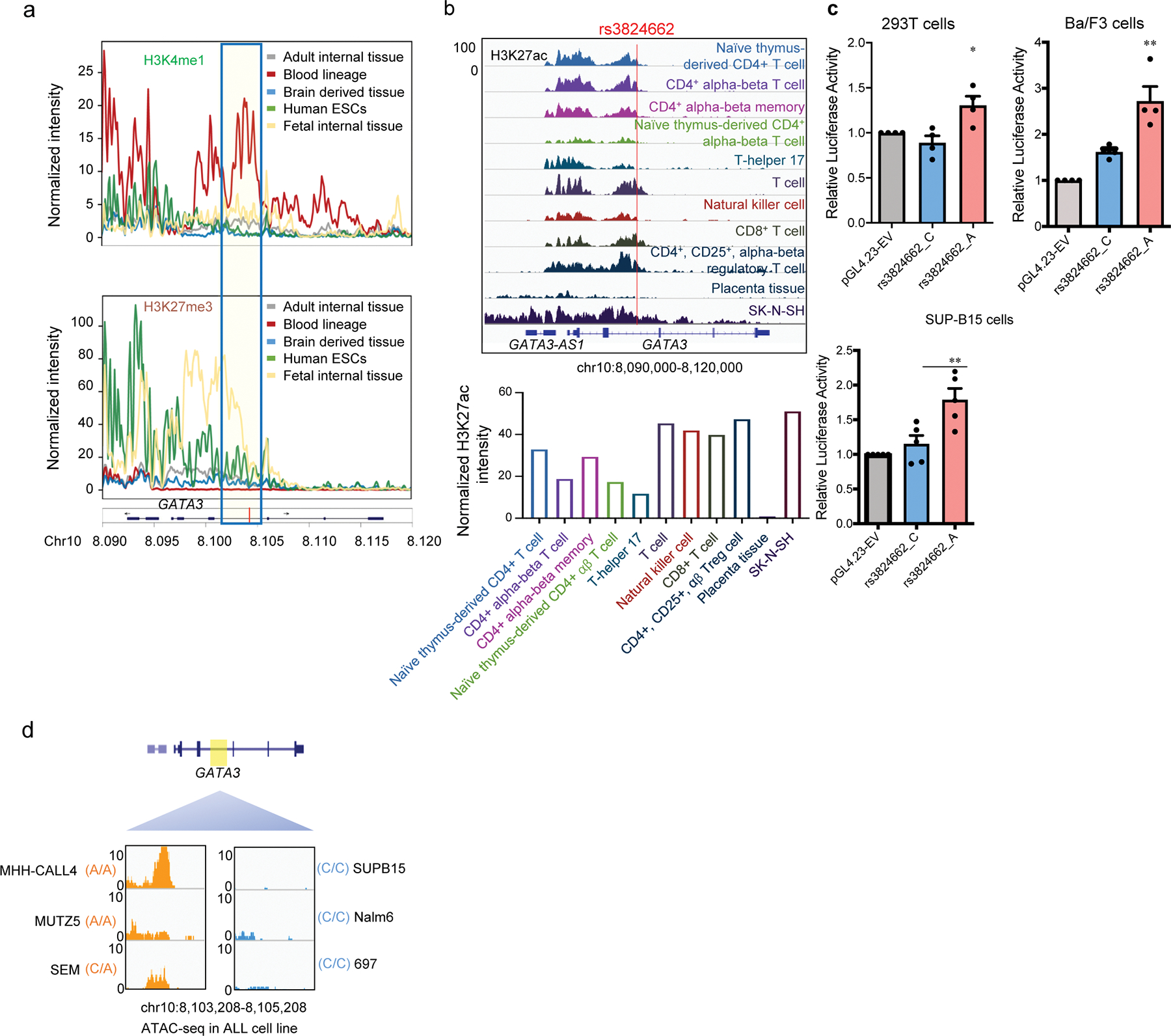
a, Normalized intensity of H3K4me1 and H3K27me3 signals at the GATA3 locus in 42 human tissues from the ROADMAP EPIGENOMICS data. Blue box indicates region encompassing rs3824662. b, H3K27ac signal of the rs3824662 locus in T cells based on ENCODE dataset. Up panel, genome browser snapshot for H3K27ac ChIP-seq signal of the rs3824662 locus in T cells. Bottom panel, the H2K27ac signal intensity of rs3824662 locus (+/− 500 bp) in T cells. c, Luciferase reporter activity comparing enhancer activities of the genomic fragments with either the rs3824662 A allele or wildtype C allele in human 293T cell(n=4), mouse Ba/F3 cell (n=4), and human ALL cell line SUP-B15(n=4). (Two-sided unpaired t-test: p value = 0.03599 for 293T; p value =0.0138 for Ba/F3; p value =0.0136 for SUP-B15). Data are presented as mean values +/− SEM. d, Open chromatin status at the rs3824662 locus (determined using ATAC-seq) in ALL cell lines representative of different molecular subtypes. The window represents a 2kb region flanking rs3824662. MHH-CALL4 and MUZT5 are CRLF2-rearranged with the A/A genotype at rs3824662; SEM is KMT2A-rearranged and with the C/A genotype, and the other three ALL cell lines have wildtype C/C genotype (SUPB15 is BCR-ABL1 ALL, Nalm6 is DUX4-rearranged, and 697 is TCF3-PBX1 ALL).
