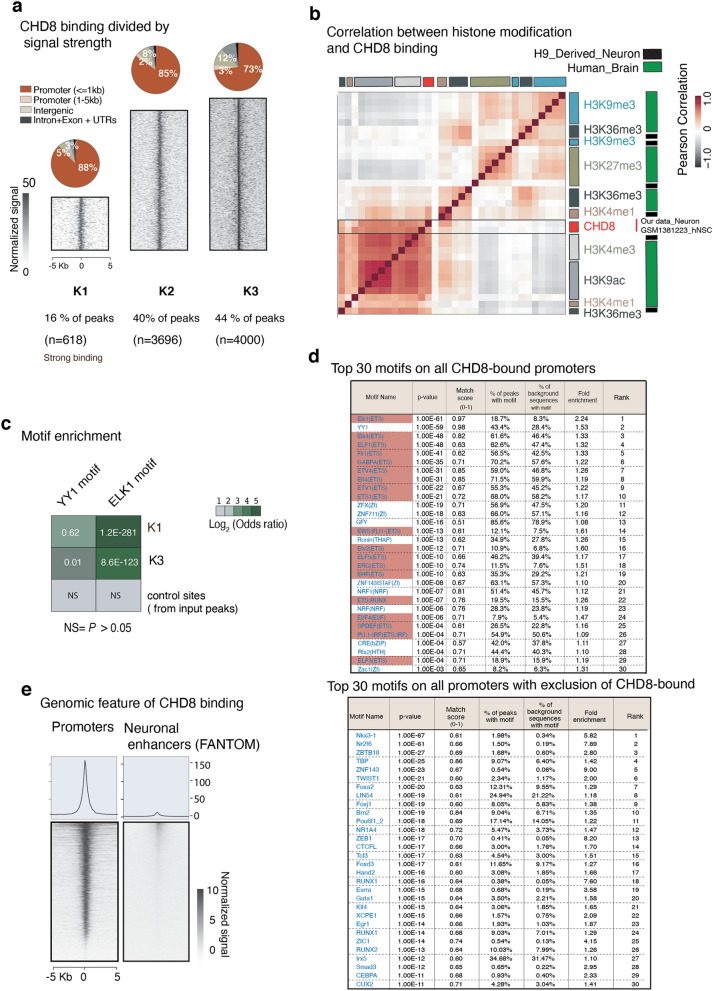
Figure 12.
CHD8 binding and enrichment at chromatin regions with distinct histone modification in neurons. (a) K-means clustering ChIP-seq signal into three groups recognize the most specific targets of CHD8 binding at promoters. Genomic annotation and functional classification are separately assigned for each group. (b) Pearson`s correlation of histone modification obtained from H9 derived human neurons (ENCODE`s histone ChIP-seq data) and our CHD8 ChIP-seq signal at CHD8 binding sites (K1 + K2 + K3). (c) The odds ratio and the calculations of motif enrichment at strong CHD8 binding sites (K1), the weak binding sites (K3), and the control sites (randomly shuffled peaks from input). Sites (K1, K2, and K3) taken from clustering analysis of peaks among CHD8 binding sites-the top heatmap Fig. 13b). (d) Top 30 motifs enriched at the prompters (± 5 Kb) of 3000 CHD8-bound target genes. The ETS factor motifs are highlighted in red. (e) Heatmap of CHD8 binding signal at promoters of neural genes and enhancers (FANTOM database)45.