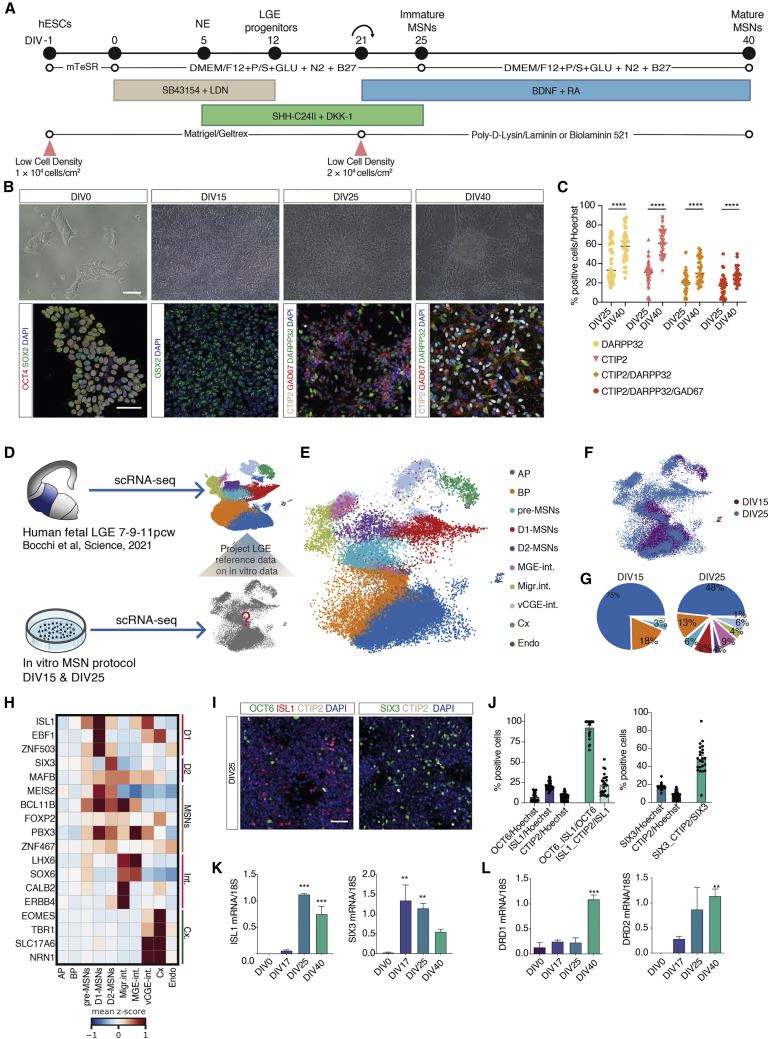
Figure 1.
MSN protocol generates cellular diversity of the human fetal striatum
(A) Schematic illustration of the protocol to obtain rapid induction of MSNs.
(B) Phase contrast images of H9 hES cells and immunofluorescent staining. Scale bars, 100 and 50 μm respectively.
(C) Quantification of the percentage of DARPP32+, CTIP2+, and GAD67+ cells. n = 9–11 independent biological replicates; error bars represent ± SEM.
(D) Overview of how cells of the human fetal LGE12 are used to classify different cell types in vitro.
(E) UMAP plot of 45,727 single cells derived from DIV15 and 25 color coded by cell type. AP, apical progenitors; BP, basal progenitors; MGE int., MGE interneurons; Migr. int., migrating interneurons; v.CGE int., ventral caudal ganglionic eminence interneurons; Cx, neocortical neurons; Endo., endothelial cells.
(F) UMAP plot of single cells color-coded by DIV.
(G) Pie charts of cell types at DIV15 and 25.
(H) Heatmap of expression values of canonical markers genes averaged by cell type.
(I) Immunofluorescent staining for D1- and D2-MSNs markers. Scale bars, 50 μm.
(J) Quantification of the percentage of D1- and D2-MSNs markers at DIV25 of differentiation. n = 3 independent biological replicates; error bars represent ± SEM.
(K and L) qRT-PCR analysis of ISL1 and SIX3 (K) and DRD1 and DRD2 (L) during differentiation. n = 3 independent differentiation experiments; error bars represent ± SEM; one-way ANOVA, Tukey’s multiple comparison test; ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figure S1.