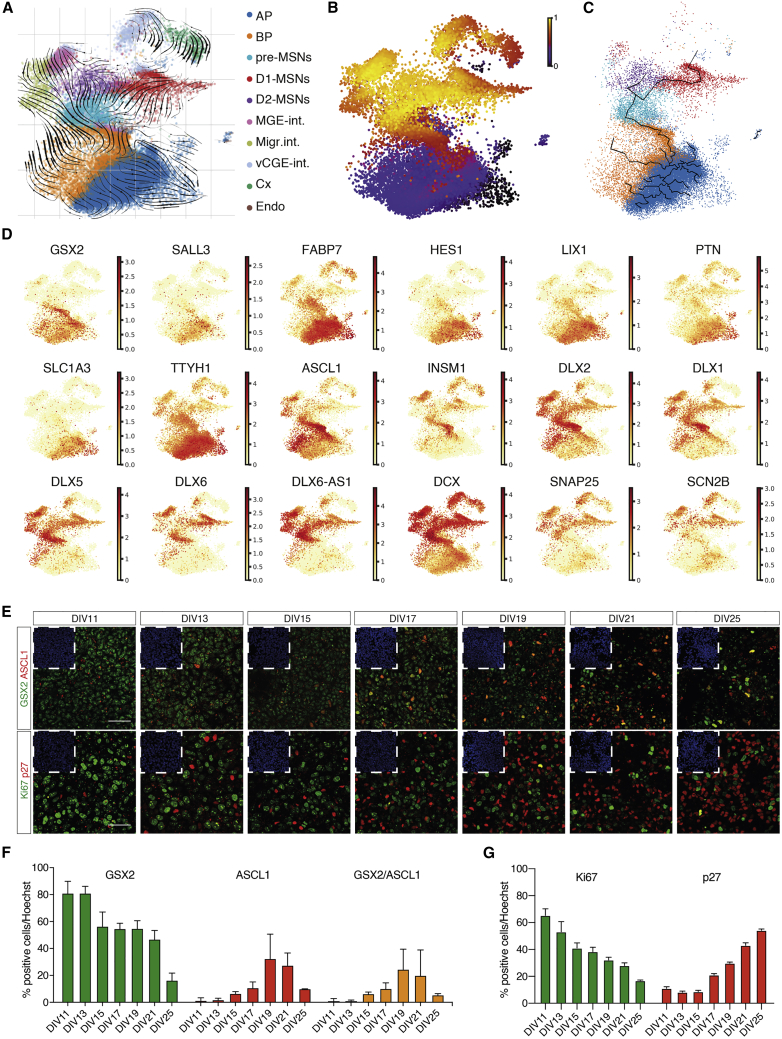
Figure 3.
MSNs are generated following the same trajectory identified in the human fetal LGE lineage
(A) Velocity estimates projected onto the UMAP plot.
(B) UMAP showing cells colored by velocity pseudotime.
(C) Trajectory predicted by Monocle.
(D) Gene expression levels plotted on the UMAP plot for specific maturation markers.
(E) Immunofluorescent staining for GSX2/ASCL1 and Ki67/p27. Top left, DAPI inset. Scale bars, 50 μm.
(F) Quantification of the proportion of GSX2, ASCL1, and GSX2+/ASCL1+ cells. n = 3 independent biological replicates; error bars represent ± SEM.
(G) Quantification of the percentage of Ki67+ and p27+ cells. n = 3 independent biological replicates; error bars represent ± SEM.